Abstract
Alzheimer disease (AD) is a leading cause of dementia in elderly patients who continue to live between 3 and 11 years of diagnosis. A steep rise in AD incidents is observed in the elderly population in East-Asian countries. The disease progresses through several changes, including memory loss, behavioural issues, and cognitive impairment. The etiology of AD is hard to determine because of its complex nature. The whole exome sequences of late-onset AD (LOAD) patients of Korean origin are investigated to identify rare genetic variants that may influence the complex disorder. Computational annotation was performed to assess the function of candidate variants in LOAD. The in silico pathogenicity prediction tools such as SIFT, Polyphen-2, Mutation Taster, CADD, LRT, PROVEAN, DANN, VEST3, fathmm-MKL, GERP + + , SiPhy, phastCons, and phyloP identified around 17 genes harbouring deleterious variants. The variants in the ALDH3A2 and RAD54B genes were pathogenic, while in 15 other genes were predicted to be variants of unknown significance. These variants can be potential risk candidates contributing to AD. In silico computational techniques such as molecular docking, molecular dynamic simulation and steered molecular dynamics were carried out to understand the structural insights of RAD54B with ATP. The simulation of mutant (T459N) RAD54B with ATP revealed reduced binding strength of ATP at its binding site. In addition, lower binding free energy was observed when compared to the wild-type RAD54B. Our study shows that the identified uncommon variants are linked to AD and could be probable predisposing genetic factors of LOAD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Alzheimer disease (AD) is a chronic, progressive neurodegenerative disorder. AD is characterized by the accumulation of extracellular neurotic beta-amyloid plaques and intracellular neurofibrillary tangles composed of hyper-phosphorylated Tau protein leading to neuronal death and cerebral atrophy (Ballard et al. 2011; Harris 2012; Holtzman et al. 2011). The disorder is associated with cognitive dysfunctions and psychiatric and behavioural disturbances. AD significantly contributes to dementia, affecting around 35 million people worldwide. Based on the onset age, AD is classified as early-onset (age < 65 years) and late-onset (age > = 65 years). Around 10% of AD patients are diagnosed with early-onset AD (EOAD) (Bekris et al. 2010). Studies have documented that the incidence rates for familial dementia and LOAD are significantly higher than population-based estimates. LOAD encompasses complex aetiology with a heritability rate of 58–79% (Vardarajan et al. 2014; Awada 2015).
The prevalence and incidence of AD indicated age as the most influential known risk factor. Early efforts in understanding AD were mainly focused on EOAD. These studies were centred on large multi-generation families' harbouring clear autosomal dominant patterns of inheritance related to mutations in genes that alter amyloid-beta (Aβ) protein production, aggregation, or clearance. EOAD patients were often associated with 3 common genes; Amyloid Beta Precursor Protein (APP) and Presenilin genes (PS1 and PS2), which occur in half of the EOAD cases. The genetic basis of LOAD is more complex, with susceptibility likely conferred by various common but less penetrant genetic factors, such as apolipoprotein E (APOE) alleles, interacting with environmental and epigenetic factors. Although substantial evidence indicates genetic factors as a key player, APOE-4, the only identified LOAD gene seems to act as a primary modifier at the age of onset and in patients with onset before age 70 years (Blacker and Tanzi 1998) (Bekris et al. 2010) (Bellenguez et al. 2019). The APOE-4 allele was overrepresented in both AD divisions. APOE-4/4 homozygotes act as a risk factor in LOAD cases.
Apart from complex genetics, other risk factors contribute to the difficulty in identifying LOAD genes. i) The base rate of LOAD cases is high and increases steeply with age. Hence clustering among families can occur due to chance alone, and several sources of the disease may co-occur in the same family. ii) LOAD occurs at the end of life span, and many individuals do not survive to the age of risk. iii) Elderly patients are prone to other sources of cognitive decline, diluting the power of genetic studies with individuals carrying the disease but are not actual gene carriers (phenocopies) (Blacker and Tanzi 1998) (Andrade-Guerrero et al. 2023). An early diagnosis of the disease is highly crucial for effective treatment. An early diagnosis helps the affected individuals to explore and benefit from drug and non-drug treatment available. An early diagnosis opens prospects for participating in wide range of clinical trials, leading to advancing research and providing medical benefits. The current treatment strategies mainly focus on reliving and delaying the progression of the symptoms.
Large-scale genome-wide association studies (GWAS) and meta-analyses of the GWAS have identified more than 30 different LOAD-susceptible loci, focusing on the European population (Karch and Goate 2015). These GWAS hits documented polymorphisms, mostly in intronic and intergenic regions. In addition, whole-exome microarray and whole-exome sequencing (WES) also contributed to identifying rare and novel variants. Recently, whole exome sequencing techniques have been utilized to understand the mutational mechanisms in several diseases, including cancer (Kumar et al. 2022). These advanced techniques also help to comprehend the structural mechanisms disrupted upon mutations using computer simulations and docking methods (Udhaya Kumar et al. 2022) (Kumar and Priya Doss 2021) (Tayubi et al. 2022).
The genetic diversity existing among different ethnic groups may have an effect on genetic factors in the pathogenesis of AD. AD genetic and clinical sucecptibility profile seems to be different with different ethnic groups (Al-Thani et al. 2021). Most of the genetic studies on AD were based on European cohorts, implying the absence of ethnic diversity data in genetic research. Including genetic studies on other populations, including East Asians, can lead to exhaustive genetic details of AD pathogenesis (Miyashita et al., 2022). The current work utilizes the distinct genetic profiles of Koreans with AD to discover LOAD-associated genes and variants. Higher AD occurrences are observed among older Koreans in East-Asian populations (Jang et al. 2021). Whole-exome sequencing analysis of post-mortem hippocampal regions from AD patients and age-matched healthy controls of Korean ethnicity is performed to identify novel LOAD risk genes.
Materials and methods
Data retrieval
The exome sequences of LOAD-affected individuals and control samples were retrieved from NCBI SRA (Accession: PRJNA532465) (Leinonen et al. 2011). Post-mortem hippocampal regions of 52 AD and 11 age-matched control samples were retrieved. About 37 LOAD samples were selected based on the disease onset (age ≥ 65 years). 15 samples were from EOAD cases (age < 65 years) and used to verify rare variants. The disease stage, ethnicity, and gender were also verified for the samples (Table 1). The selected samples were chosen for further analysis.
Data processing
The quality control of the raw paired-end exome sequences was carried out using FastQC. Following quality control, high-quality reads were mapped to human genome build GRCh38 using Burrows-Wheeler Aligner (Li and Durbin 2009). The mapped reads were duplicate marked, and base quality score recalibration was carried out using Picard and GATK suite. GATK suite was employed to identify variants. The variants were annotated using Annotate Variation (ANNOVAR) tool (Wang et al. 2010). Non-synonymous exonic variants with Minor Allele Frequencies (MAF < 0.01) were prioritized to identify rare variants. Only coding variants were selected. Variant pathogenicity was assessed using SIFT, Polyphen-2, Mutation Taster, CADD, LRT, PROVEAN, DANN, VEST3, fathmm-MKL, GERP, SiPhy, phastCons, and phyloP tools (Doss and Zayed 2017; Liu et al. 2020). The final candidate variants were prioritized by utilizing UKBiobank PheWeb, GeneCards (Safran et al. 2010), OMIM (Amberger et al. 2015), and ACMG guidelines (Harrison et al. 2019).
Docking
Using the molecular docking approach, users can theoretically screen a library of chemicals and forecast the most potent binding sites using a variety of scoring algorithm (Agrahari et al. 2018a, 2019; Ali et al. 2017). Using Autodock Vina docking software, the docking analysis of the ligand ATP with RAD54B was performed. The predicted active site was the primary target for docking the protein–ligand complex. The anticipated active site of RAD54B was then docked with arylidenes ATP using AutodockVina (Trott and Olson 2010). The pose with the highest AutodockVina dock score was then chosen for molecular dynamics simulation studies.
Protein–ligand dynamic simulation (MDS)
Selected Protein–Ligand complexes from docking data were subjected to MDS using Gromacs-2019 (Agrahari et al. 2018b; Abraham et al. 2015). We obtained the chosen ligand topology by downloading it from the PRODRG website. Utilizing the steepest descent technique, we setup the system and reduced the vacuum for 1500 steps (Petrova and Solov'ev 1997). The structures in a cubic periodic box of 0.5 nm were solvated using the simple point charge (SPC) water model. It was, therefore, sufficient to sustain the complex system with a salt concentration of 0.15 M by introducing an adequate amount of Na+ and Cl− counter ions. The system setup was covered based on a previously published work. The ensemble underwent a final simulation run of 100 ns following the NPT equilibration stage. The trajectory was examined using various GROMACS analytic techniques as a last step. The RMSD, RMSF, Rg, solvent accessible surface area (SASA), and intramolecular H-bonds between our protein molecules were calculated for the wild and all mutant proteins, respectively, using the gmx rms, gmx rmsf, gmx gyrate, gmx sasa, and gmx hbond. Molecular Mechanics Poisson-Boltzmann Surface Area (MM-PBSA) was employed to comprehend a substrate's binding free energy (ΔG binding) with a protein throughout the simulation. The GROMACS function g_mmpbsa was utilized to estimate the ΔG binding. Within the final 1000 frames, in the last 50 ns, computation ΔG produced our results (Homeyer and Gohlke 2012).
Steered molecular dynamics
We ran a brief MD run on each system to establish equilibrated structures before running the SMD simulation (Izrailev et al. 1999). The Alphafold structure of RAD54B uploaded to UniProt served as the basis for the MD simulations. The GROMOS54A7 force field was used to run the simulations for both the wild and mutant RAD54B-ATP systems (Huang et al. 2011). GROMOS54A7 force field compatible parameters for the ATP molecule were retrieved from the PRODRG server (Schüttelkopf and van Aalten 2004). The structures were subjected to vacuum energy minimization before using the steepest descent algorithm. With the help of the SPC model, water molecules extended 20 Å from the protein across all sides of the cubic box in which the structures were solvated. Using the steepest descent algorithm, an appropriate number of ions was added to maintain a salt concentration of 0.15 M. The systems were energy-minimized for 5000 steps. The systems were first brought into equilibrium in the NVT ensemble and then in the NPT ensemble for 100 ps each. A V-rescale thermostat 37 with a coupling constant of 0.1 ps was used to maintain the physiological temperature of 310 K, and a Parinello-Rahman barostat with isotropic pressure coupling was used to maintain the pressure at 1 bar. The long-range electrostatic interactions were handled by the particle mesh Ewald (PME) sum with a cutoff of 1.0 nm (Essmann et al. 1995). Periodic boundary conditions and a threshold radius of 1.0 nm were employed for van der Waals interactions.
Using the LINCS algorithm, all hydrogen atom bonds were restricted (Hess et al. 1997). The final equilibrated structure from the NPT equilibration step was considered the initial structure for performing Steered Molecular Dynamics (SMD) Simulation Studies. Various biomolecular systems have been evaluated using constant-velocity SMD simulation, and encouraging correlations with experimental results are reported. To investigate the ATP molecule's binding affinity at the ATP binding site, separate MD simulations were run on each system. Using constant-velocity SMD simulations, ATP was pulled from the ATP binding site in each system. The centre of mass of the steered group, or the ATP tail region, and the centre of mass of the protein residues constituting the ATP binding pocket were used to define the pulling vector. The constant velocity simulations used a force constant of 1000 kJ mol-1 nm-2. ATP was drawn with a 5 nm/ns pulling velocity for each system. We emphasize that the systems are frequently severely perturbed when greater pulling rates are used in SMD, and some details are less likely to be recorded at such higher pulling rates. The increased pulling rates can also impair the protein's normal elastic response. The pulling velocity was selected to achieve the ideal balance of precision and computing speed.
Results and discussion
The exomes of individuals affected with LOAD were compared with healthy controls. Around 85,000 variants per sample qualifying the initial quality control filters were investigated further. The filtering of variants for synonymous variants and known LOAD risk genes (ABI3, ABCA7, ADAM17, AKAP9, IGHG3, BIN1, CASS4, CD33, CD2AP, CELF1, CLU, CR1, DSG2, EPHA1, FERMUTANT2, HLA-DRB5-DBR1, INPP5D, MS4A, MEF2C, NME8, PICALM, PLD3, PLCG2, PTK2B, SLC24H4-RIN3, SORL1, UNC5C, and ZCWPW1) resulted in the identification of 1531 variants.
Rare variants identification in LOAD samples
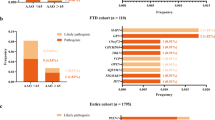
Filtering variants with MAF < 0.01 resulted in 1151 hits. Variants from 21 genes GJA8, CHRNB3, PGK2, TGM2, GDF9, CCR10, ALDH3A2, CLDN3, GK2, GSR, MUTANTHFS; ST20-MUTANTHFS, CHRNA1, RAD54B, GDA, TMLHE, MIXL1, CPXM1, WNT10A, KLC4, WFDC2, and MAGI1 were predicted to be damaging to the protein by SIFT, Polyphen-2, Mutation Taster, CADD, LRT, PROVEAN, DANN, VEST3, fathmm-MKL, and SiPhy. These variants were also predicted to be conserved by GERP, phastCons, and phyloP. Systems-level investigation of these variants was carried out using OMIM and ACMG guidelines. Significant associations were observed between 9 genes (GJA8, CHRNB3, GDF9, ALDH3A2, GSR, CHRNA1, GDA, KLC4, and MAGI1) and the disease phenotype using UKBiobank PheWeb. The presence of these variants was checked in control and EOAD samples. The comparison showed that variants from 17 genes were absent in control or EOAD cases (Tables 2 & 3). The genes ALDH3A2 and RAD54B harboured likely pathogenic variants, and the remaining 15 genes contained variants of unknown significance. The genes predicted to be deleterious were further assessed for their involvement in the disease.
Functional insight into genes associated with LOAD
Pathogenic variants
The variants from two genes, ALDH3A2 and RAD54B, were predicted to be deleterious and pathogenic by the in silico tools.
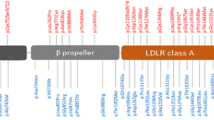
ALDH3A2 reported two single nucleotide variants (SNV) at exon 7. The gene product is an NAD + oxidoreductase enzyme complex component responsible for oxidizing fatty alcohol to a fatty acid. Mouse knockout studies of the gene showed several abnormalities corresponding to behavioural traits correlating with movement instability and anxiety issues observed in AD patients. The SNV observed at exon 7 of the ALDH3A2 gene in our sample correlates with the point mutation associated with Sjogren-Larsson Syndrome (SLS). The mutation causes C to T exchange at the nucleotide position 943 in the cDNA leading to the replacement of highly conserved proline to serine (De Laurenzi et al. 1997; Kanetake et al. 2019).
RAD54B reported two SNVs at exon 6 and exon 8. The gene is a member of the helicase superfamily involved in recombination and DNA repair. DNA damage is one of the critical pathological causes of AD, as DNA damage accumulation is noted in patients' brains. Defects in DNA damage and repair enzymes such as RAD54B may facilitate the disease pathogenesis (Murzik et al. 2008; Lin et al. 2020).
Uncertain significant variants
Around 15 genes (CHRNB3, GDF9, CCR10, CLDN3, GK2, GSR, MUTANTHFS; ST20-MUTANTHFS, GDA, TMLHE, MIXL1, CPXM1, WNT10A, KLC4, WFDC2, and MAGI1) with uncertain significance variants and severe consequences were obtained (Tables 4 and 5).
CHRNB3 reported two SNVs in exons 5 and 6. The gene codes for the neuronal nicotinic acetylcholine receptor (nAChR) component. The variant associated with CHRNB3 may potentially affect the regulation of nAChRs leading to the disruption of transmitter release, neuronal integration, and cell excitability, as documented in many neurological disorders (Hogg et al. 2003; Abu-Amero et al. 2015). CCR10 reported an SNV in exon 2. CCR10 is a chemokine receptor expressed in astrocytes. Mutation in the gene may alter its ligand binding, inducing immune cascade disturbances in the Central Nervous System (Liu et al. 2014). CLDN3 also reported an SNV in exon 1. The gene codes for a component of tight junction strands. CLDN3 is known to localize in the brain endothelial cells. Any genetic changes in the gene may introduce a breakdown in the blood–brain barrier associated with the endothelial cells (Romanitan et al. 2010). GDA/Cypin reported around seven SNV and observed among four samples in our dataset. The gene is localized in dendrites, increasing branching and promoting microtubule assembly. Patients with AD show fewer branches of neurons within the hippocampus, potentially reflecting the loss of learning and memory (Arikkath 2012). TMLHE reported two SNVs in exon 6. The gene product is the first enzyme in the carnitine biosynthesis pathway. Carnitine contributes to neuroprotective, neuromodulatory, and neurotrophic functions. Mutations in the gene may affect the carnitine pathway. Loss of function of TMLHE is also shown to be associated with autism spectrum disorders (Nałecz et al. 2004; Virmani and Binienda 2004; Nava et al. 2012).
KLC4 reported five SNVs in exons 7, 8, and 9. Studies on mutant KLC4 zebrafish revealed that it is crucial for peripheral sensory axon branching and proper arborization. The study also showed altered microtubule dynamics. An increased anxiety-like behaviour was also observed, indicating the role of KLC4 in neural circuits (Haynes et al. 2022). MAGI1 reported an SNV in exon 6. The gene plays a significant role in the organization of membrane proteins and cytoskeletons by transmitting signals pertaining to cell–cell or cell–matrix interactions. Mutations in MAGI1 may result in the perturbation of these cell–cell signalling (Hammad et al. 2016). WNT10A reported an SNV in exon 3. WNT10A−/− knockout mice exhibited spatial memory impairment and anxiety-like behaviour (Zhang et al. 2022). WNT10A deficiency was proven to cause hippocampal neuro-degeneration in mice indicating similar effects in AD patients. WFDC2 reported an SNV in exon 2. Human epididymis protein 4 (HE4), encoded by the WFDC2 gene, is a secretory protein expressed in human epididymis and an important biomarker for ovarian epithelial cancer (James et al. 2018). Studies on serum levels of HE4 revealed it as a sensitive biomarker for the early recognition of the cognitive decline in patients suffering from diabetes mellitus (Bai et al. 2020). As observed in AD patients, impairment in the WFDC2 gene function may lead to cognitive decline.
One of the limitations of the current study is that no literature support was found to assess the function and involvement of other genes (GDF9, GK2, GSR, MUTANTHFS; ST20-MUTANTHFS, MIXL1 and CPXM1) in AD or other neurological disorders.
Molecular dynamics (MDS)
Through 100-ns MD simulations, we carried out all-atom MDS to examine the consequences of the complex mutation T459N on the structural integrity of the RAD54B protein [Wild type with ATP (Wild-ATP) and mutant type [T459N] with ATP (Mutant-ATP)].
After 20 ns of observation, we discovered that these trajectory motions grew steadier. As a result, the second half of the trajectory was considered for additional investigation. The mutant complex's RMSD measurements did not reveal appreciable variations in these outcomes. The RMSD of Wild-AT and Mutant-ATP complex is illustrated in (Fig. 1). The RMSD value has produced a consistent trajectory, offering a good foundation for further research. The Wild-ATP, Mutant-ATP, and average RMSD values were 0.2554 and 0.2802.
To evaluate and fully grasp the effects of the Wild-AT and Mutant-ATP complex on the flexible areas of the RAD54B, the Ca residue (RMSF) was determined from its time-averaged stance. The RMSF describes that the residue backbone adopts a higher level of fluctuation in Mutant-ATP than in the Wild-ATP complex. It is hypothesized that the mutation T459N alters how ATP binds to the protein and increases the backbone's flexibility. The RMSF information of Wild-ATP and Mutant-ATP complexes is illustrated in (Fig. 2).
The mass-weighted root mean square distance of atoms from their centers of mass can be used to characterize the radius of gyration (Lobanov et al. 2008). The Rg figure shows the competency and form folding of the entire RAD54B structure at various times during the trajectory (Fig. 3). Throughout the simulation, the Wild-ATP complex exhibited a nearly similar Rg value of which the Mutant-ATP complex showed a higher deviation of Rg. As a result, the mutant protein has become more compact, resulting in a slower folding rate than the Wild-ATP.
To assess the hydrophobic core's compactness, the SASA change was studied. The change of SASA of the Wild-ATP and Mutant-ATP complexes with time is shown in (Fig. 4). Both complexes exhibited a nearly similar SASA value throughout the simulation.
The H-bonds numbers formed between RAD54B and ATP during the MDS were also evaluated. The H-bond profile was varied, fluctuating from 0 to 6 with a median of 3 H-bonds in the Wild-ATP and Mutant-ATP complexes (Fig. 5). The average values for the WT-ATP and MT-ATP simulations are provided in Table 5.
Steered molecular dynamics (SMD)
On-time scales attainable by molecular dynamics simulations steered molecular dynamics (SMD) causes ligands to unbind from their biomolecules and change in conformation. A system is subjected to time-dependent external influences, and the system's responses are studied. In the present work, SMD simulations pulled ATP along its unbinding path. SMD simulations were run using the stiff spring constant to pull the ATP molecule from its binding site on the RAD54B protein. In the case of the Wild-ATP complex, the force value increased along the time evolution of the pulling simulation during the initial unbinding phase of ATP (0–600 ps). ATP was released from the binding pocket with a peak rupture force of 925.27 kJ mol−1 nm−1. However, in the Mutant-ATP complex, the ATP molecule was found to be released from the ATP binding pocket with a similar peak rupture force value of 978.37 kJ mol−1 nm−1. The forces start to decline after reaching a maximum, indicating disruption of strong non-bonded interactions between ATP and the lining residues of the ATP binding pocket. The force profile of the Wild-ATP complex was relatively flat after 700 ps, indicating only modest interaction in the dissociation path. However, the force plot of the Mutant-ATP complex became flat after 400 ps. Thus, the ATP molecule is difficult to pull out of the binding pocket in the case of wild protein, as strong non-bonded interactions exist between ATP and the pocket-lining residues. This fact is reflected in the force plot of the Wild-ATP complex, where a force of 900 kJ mol−1 nm−1 was applied in large timeframes (350-600 ps) to rupture all interactions and release the molecule. This was not observed in the case of the Mutant-ATP complex as the pull force was able to rupture the interactions between ATP and the protein in a very short time frame (300-340 ps), indicating that the ATP molecule weakly bound in the mutant protein, thereby capable of being extracted very quickly from the complex. The Wild-ATP and Mutant-ATP complexes of the SMD graph are illustrated in (Fig. 6).
Wild-ATP and mutant-ATP complexes' binding affinities were measured. Within the active site, we looked at the differential binding capacity. (Fig. 6) compares the binding strength of Wild-ATP and Mutant-ATP complexes examined via the MM-PBSA method. We determined residue-level contributions to the interaction energy throughout a steady simulation trajectory.
(Fig. 7) demonstrates that the binding energy of Wild-ATP in the active center pocket was found to be -10.5416 kcal/mol, while the Mutant-ATP procured binding energy of -3.401 kcal/mol. The MM-PBSA suggested that Wild-ATP exhibited significant binding energy in the active binding pocket. They show that Wild-ATP interacted with the active site pocket more favourably than Mutant-ATP (Table 5). Hence, the above computational analyses exhibited the importance of wild RAD54B compared to the mutant RAD54B. These differences would make the mutant RAD54B either disrupt its functional role due to mutation or could influence the pathway that the RAD54B involved. However, a wider population and biochemical techniques are required to validate the RAD54B mutations in AD patients.
Conclusion
Identifying genetic modifiers is crucial in understanding their significant contribution to the disease's pathogenesis. In the current study, 37 whole exome sequences of LOAD samples were investigated to identify rare variants specific to the East-Asian population. Around 17 genes with potentially deleterious variants not previously studied in LOAD cases were identified. These rare variants are reported for the first time in individuals with LOAD from our study. Association signals that GWAS previously discovered with common and primarily non-functional variants cannot be explained by rare variants. However, our study findings explicitly showed the structural mechanisms of RAD54B mutation and also, more target-based biological experiments should be implemented to learn more about how these genes contribute to AD pathogenesis. Nevertheless, the identified variants were not previously linked to AD, highlighting the capacity of the whole-exome sequencing method to find uncommon variations linked to AD. These rare variants could be considered novel predisposing genetic factors for LOAD and might increase neuro-degeneration.
Data availability
Data sharing is not applicable- no new data is generated, or the article describes entirely theoretical research.
References
Abraham MJ, Murtola T, Schulz R et al (2015) GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2:19–25. https://doi.org/10.1016/j.softx.2015.06.001
Abu-Amero KK, Kondkar A, Hellani AM et al (2015) Nicotinic Receptor Mutation in a Mildly Dysmorphic Girl with Duane Retraction Syndrome. Ophthalmic Genet 36:99–104. https://doi.org/10.3109/13816810.2013.835431
Agrahari AK, Kumar A, Silva R, Zayed H, Doss GPC (2018a) Substitution impact of highly conserved arginine residue at position 75 in GJB1 gene in association with X-linked Charcot-Marie-tooth disease: A computational study. J Theor Biol 437:305–317
Agrahari AK, Muskan M, Doss CGP, Siva R, Zayed H (2018b) Computational insights of K1444N substitution in GAP-related domain of NF1 gene associated with neurofibromatosis type 1 disease: a molecular modeling and dynamics approach. Metab Brain Dis 33:1443–1457
Agrahari AK, Doss GPC, Siva R, Magesh R, Zayed H (2019) Molecular insights of the G2019S substitution in LRRK2 kinase domain associated with Parkinson’s disease: A molecular dynamics simulation approach. J Theor Biol 469:163–171
Ali SK, Sneha P, Priyadharshini Christy J, Zayed H, Doss CGP (2017) Molecular dynamics-based analyses of the structural instability and secondary structure of the fibrinogen gamma chain protein with the D356V mutation. J Biomol Struct Dyn 35:2714–2724
Al-Thani HF, Ahmad MN, Younes S, Zayed H (2021) Genetic Variants Associated With Alzheimer Disease in the 22 Arab Countries: A Systematic Review. Alzheimer Dis Assoc Disord 35:178–186
Amberger JS, Bocchini CA, Schiettecatte F et al (2015) OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res 43:D789-798. https://doi.org/10.1093/nar/gku1205
Andrade-Guerrero J, Santiago-Balmaseda A, Jeronimo-Aguilar P, Vargas-Rodríguez I, Cadena-Suárez AR, Sánchez-Garibay C et al (2023) Alzheimer’s Disease: An Updated Overview of Its Genetics. Int J Mol Sci 24(4):3754
Arikkath J (2012) Molecular mechanisms of dendrite morphogenesis. Front Cell Neurosci 6:61. https://doi.org/10.3389/fncel.2012.00061
Awada AA (2015) Early and late-onset Alzheimer’s disease: What are the differences? J Neurosci Rural Pract 6:455–456. https://doi.org/10.4103/0976-3147.154581
Bai F, Li T, Li B, Li X, Zhu L (2020) Serum Human Epididymis Protein 4 Level is Associated with Cognitive Function in Patients with Diabetes Mellitus. Diabetes Metab Syndr Obes 13:3919–3924
Ballard C, Gauthier S, Corbett A et al (2011) Alzheimer’s disease. The Lancet 377:1019–1031. https://doi.org/10.1016/S0140-6736(10)61349-9
Bekris LM, Yu C-E, Bird TD, Tsuang DW (2010) Genetics of Alzheimer disease. J Geriatr Psychiatry Neurol 23:213–227. https://doi.org/10.1177/0891988710383571
Bellenguez C, Grenier-Boley B, Lambert JC (2020) Genetics of Alzheimer’s disease: where we are, and where we are going. Curr Opin Neurobiol 61:40–48
Blacker D, Tanzi RE (1998) The genetics of Alzheimer disease: current status and future prospects. Arch Neurol 55(3):294–296
De Laurenzi V, Rogers GR, Tarcsa E et al (1997) Sjögren-Larsson syndrome is caused by a common mutation in northern European and Swedish patients. J Invest Dermatol 109:79–83. https://doi.org/10.1111/1523-1747.ep12276622
Doss GPC, Zayed H (2017) Comparative computational assessment of the pathogenicity of mutations in the Aspartoacylase enzyme. Metab Brain Dis 32:2105–2118
Essmann U, Perera L, Berkowitz ML et al (1995) A smooth particle mesh Ewald method. J Chem Phys 103:8577–8593. https://doi.org/10.1063/1.470117
Hammad MM, Dunn HA, Ferguson SSG (2016) MAGI Proteins Regulate the Trafficking and Signaling of Corticotropin-Releasing Factor Receptor 1 via a Compensatory Mechanism. J Mol Signal 11:5. https://doi.org/10.5334/1750-2187-11-5
Harris JR (2012) Protein Aggregation and Fibrillogenesis in Cerebral and Systemic Amyloid Disease. Springer Science & Business Media
Harrison SM, Biesecker LG, Rehm HL (2019) Overview of Specifications to the ACMG/AMP Variant Interpretation Guidelines. Curr Protoc Hum Genet 103:e93. https://doi.org/10.1002/cphg.93
Haynes EM, Burnett KH, He J et al (2022) KLC4 shapes axon arbors during development and mediates adult behavior. ELife 11:e74270. https://doi.org/10.7554/eLife.74270
Hess B, Bekker H, Berendsen HJC, Fraaije JGEM (1997) LINCS: A linear constraint solver for molecular simulations. J Comput Chem 18:1463–1472. https://doi.org/10.1002/(SICI)1096-987X(199709)18:12%3c1463::AID-JCC4%3e3.0.CO;2-H
Hogg RC, Raggenbass M, Bertrand D (2003) Nicotinic acetylcholine receptors: from structure to brain function. Rev Physiol Biochem Pharmacol 147:1–46. https://doi.org/10.1007/s10254-003-0005-1
Holtzman DM, Morris JC, Goate AM (2011) Alzheimer’s Disease: The Challenge of the Second Century. Sci Transl Med 3:77sr1-77sr1. https://doi.org/10.1126/scitranslmed.3002369
Homeyer N, Gohlke H (2012) Free Energy Calculations by the Molecular Mechanics Poisson-Boltzmann Surface Area Method. Mol Inform 31:114–122. https://doi.org/10.1002/minf.201100135
Huang W, Lin Z, van Gunsteren WF (2011) Validation of the GROMOS 54A7 Force Field with Respect to β-Peptide Folding. J Chem Theory Comput 7:1237–1243. https://doi.org/10.1021/ct100747y
Izrailev S, Stepaniants S, Isralewitz B et al (1999) Steered Molecular Dynamics. In: Deuflhard P, Hermans J, Leimkuhler B et al (eds) Computational Molecular Dynamics: Challenges, Methods, Ideas. Springer, Berlin, Heidelberg, pp 39–65
James NE, Chichester C, Ribeiro JR (2018) Beyond the Biomarker: Understanding the Diverse Roles of Human Epididymis Protein 4 in the Pathogenesis of Epithelial Ovarian Cancer. Front Oncol 8:124
Jang JW, Park JH, Kim S et al (2021) Prevalence and Incidence of Dementia in South Korea: A Nationwide Analysis of the National Health Insurance Service Senior Cohort. J Clin Neurol Seoul Korea 17:249–256. https://doi.org/10.3988/jcn.2021.17.2.249
Kanetake T, Sassa T, Nojiri K et al (2019) Neural symptoms in a gene knockout mouse model of Sjögren-Larsson syndrome are associated with a decrease in 2-hydroxygalactosylceramide. FASEB J off Publ Fed Am Soc Exp Biol 33:928–941. https://doi.org/10.1096/fj.201800291R
Karch CM, Goate AM (2015) Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol Psychiatry 77:43–51. https://doi.org/10.1016/j.biopsych.2014.05.006
Kumar SU, Priya Doss CG (2021) Computational investigation to identify potent inhibitors of the GTPase-Kirsten RAt sarcoma virus (K-Ras) mutants G12C and G12D. Comput Biol Med 139:104946. https://doi.org/10.1016/j.compbiomed.2021.104946
Kumar SU, Balasundaram A, Cathryn RH et al (2022) Whole-exome sequencing analysis of NSCLC reveals the pathogenic missense variants from cancer-associated genes. Comput Biol Med 148:105701. https://doi.org/10.1016/j.compbiomed.2022.105701
Leinonen R, Sugawara H, Shumway M, International Nucleotide Sequence Database Collaboration (2011) The sequence read archive. Nucleic Acids Res 39:D19-21. https://doi.org/10.1093/nar/gkq1019
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinforma Oxf Engl 25:1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Lin X, Kapoor A, Gu Y et al (2020) Contributions of DNA Damage to Alzheimer’s Disease. Int J Mol Sci 21:1666. https://doi.org/10.3390/ijms21051666
Liu C, Cui G, Zhu M et al (2014) Neuroinflammation in Alzheimer’s disease: chemokines produced by astrocytes and chemokine receptors. Int J Clin Exp Pathol 7:8342–8355
Liu X, Li C, Mou C et al (2020) dbNSFP v4: a comprehensive database of transcript-specific functional predictions and annotations for human nonsynonymous and splice-site SNVs. Genome Med 12:103. https://doi.org/10.1186/s13073-020-00803-9
Lobanov MYu, Bogatyreva NS, Galzitskaya OV (2008) Radius of gyration as an indicator of protein structure compactness. Mol Biol 42:623–628. https://doi.org/10.1134/S0026893308040195
Miyashita A, Kikuchi M, Hara N, Ikeuchi T (2023) Genetics of Alzheimer’s disease: an East Asian perspective. J Hum Genet 68(3):115–124
Murzik U, Hemmerich P, Weidtkamp-Peters S et al (2008) Rad54B targeting to DNA double-strand break repair sites requires complex formation with S100A11. Mol Biol Cell 19:2926–2935. https://doi.org/10.1091/mbc.e07-11-1167
Nałecz KA, Miecz D, Berezowski V, Cecchelli R (2004) Carnitine: transport and physiological functions in the brain. Mol Aspects Med 25:551–567. https://doi.org/10.1016/j.mam.2004.06.001
Nava C, Lamari F, Héron D et al (2012) Analysis of the chromosome X exome in patients with autism spectrum disorders identified novel candidate genes, including TMLHE. Transl Psychiatry 2:e179. https://doi.org/10.1038/tp.2012.102
Petrova SS, Solov’ev AD (1997) The Origin of the Method of Steepest Descent. Hist Math 24:361–375. https://doi.org/10.1006/hmat.1996.2146
Romanitan MO, Popescu BO, Spulber S et al (2010) Altered expression of claudin family proteins in Alzheimer’s disease and vascular dementia brains. J Cell Mol Med 14:1088–1100. https://doi.org/10.1111/j.1582-4934.2009.00999.x
Safran M, Dalah I, Alexander J et al (2010) GeneCards Version 3: the human gene integrator. Database J Biol Databases Curation 2010:baq020. https://doi.org/10.1093/database/baq020
Schüttelkopf AW, van Aalten DMF (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr 60:1355–1363. https://doi.org/10.1107/S0907444904011679
Tayubi IA, Kumar SU, Doss CGP (2022) Identification of potential inhibitors, conformational dynamics, and mechanistic insights into mutant Kirsten rat sarcoma virus (G13D) driven cancers. J Cell Biochem 123:1467–1480. https://doi.org/10.1002/jcb.30305
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461. https://doi.org/10.1002/jcc.21334
Udhaya Kumar S, Kamaraj B, Varghese RP et al (2022) Mutations in G6PC2 gene with increased risk for development of type 2 diabetes: Understanding via computational approach. Adv Protein Chem Struct Biol 130:351–373. https://doi.org/10.1016/bs.apcsb.2022.02.005
Vardarajan BN, Faber KM, Bird TD et al (2014) Age-specific incidence rates for dementia and Alzheimer disease in NIA-LOAD/NCRAD and EFIGA families: National Institute on Aging Genetics Initiative for Late-Onset Alzheimer Disease/National Cell Repository for Alzheimer Disease (NIA-LOAD/NCRAD) and Estudio Familiar de Influencia Genetica en Alzheimer (EFIGA). JAMA Neurol 71:315–323. https://doi.org/10.1001/jamaneurol.2013.5570
Virmani A, Binienda Z (2004) Role of carnitine esters in brain neuropathology. Mol Aspects Med 25:533–549. https://doi.org/10.1016/j.mam.2004.06.003
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:e164. https://doi.org/10.1093/nar/gkq603
Zhang JH, Tasaki T, Tsukamoto M, Wang KY, Kubo KY, Azuma K (2022) Deletion of Wnt10a Is Implicated in Hippocampal Neurodegeneration in Mice. Biomedicines 10(7):1500
Acknowledgements
Open Access funding provided by the Qatar National Library. The authors also would like to thank the management of the Vellore Institute of Technology (VIT), Vellore, Tamil Nadu, India, for providing the necessary facilities and encouragement to carry out this work.
Funding
No funding agency involved in the present study.
Author information
Authors and Affiliations
Contributions
SS, AV, UKS, MG, ITA, and GPDC were involved in the study design. SS, AV, UKS, AM, MG, ITA and GBS contributed to the analysis, data interpretation, and manuscript drafting. HZ and CGPD supervised the entire study and were involved in the study design, the acquisition, analysis, and understanding of the data, and critically reviewed the manuscript. All authors edited and approved the submitted version of the article.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that the study has no conflict of interest.
Ethics approval (include appropriate approvals or waivers)
Not Applicable.
Consent to participate (include appropriate statements)
Not Applicable.
Consent for publication (include appropriate statements)
Not Applicable.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sundarrajan, S., Venkatesan, A., Kumar S, U. et al. Exome sequence analysis of rare frequency variants in Late-Onset Alzheimer Disease. Metab Brain Dis 38, 2025–2036 (2023). https://doi.org/10.1007/s11011-023-01221-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11011-023-01221-7