Abstract
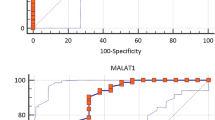
mTOR has been shown to be involved in the regulation of immune responses and differentiation of immune cells. This protein is a candidate molecule for unraveling the molecular mechanisms of autoimmune disorders such as multiple sclerosis (MS). We designed the current study to assess expression of MTOR, and four associated long non-coding RNAs (lncRNAs), namely SNHG1, SNHG3, SHNG5 and DANCR in the peripheral blood of patients with MS compared with healthy controls. Analysis of real-time PCR results has shown down-regulation of SNHG5 and DANCR in MS patients compared with controls. Sex of study participants had no significant effect on expression of either genes and the interaction of sex and disease on expression levels of all studied genes were insignificant. There was a significant negative correlation between expression levels of MTOR gene and disease duration. No other significant correlations were detected between genes expressions and clinical/demographic data. SNHG5 and DANCR transcript levels had AUC values of 0.88 and 0.68 in separation of patients with MS from healthy controls, respectively. Taken together, our study suggests participation of two mTOR-related lncRNAs, i.e. SNHG5 and DANCR in the pathophysiology of MS.



Similar content being viewed by others
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Amiri M, Mokhtari MJ, Bayat M, Safari A, Dianatpuor M, Tabrizi R, Borhani-Haghighi A (2022) Expression and diagnostic values of MIAT, H19, and NRON long non-coding RNAs in multiple sclerosis patients. Egypt J Med Hum Genet 23:1–9
Chapman NM, Chi H (2014) mTOR signaling, Tregs and immune modulation. Immunotherapy 6:1295–1311
Giacoppo S, Pollastro F, Grassi G, Bramanti P, Mazzon E (2017) Target regulation of PI3K/Akt/mTOR pathway by cannabidiol in treatment of experimental multiple sclerosis. Fitoterapia 116:77–84
Han W, Shi J, Cao J, Dong B, Guan W (2020) Latest Advances of Long Non-Coding RNA SNHG5 in Human Cancers. Onco Targets Ther 13:6393–6403
Hermida MA, Kumar JD, Leslie NR (2017) GSK3 and its interactions with the PI3K/AKT/mTOR signalling network. Adv Biol Regul 65:5–15
Jhanwar-Uniyal M, Amin AG, Cooper JB, Das K, Schmidt MH, Murali R (2017) Discrete signaling mechanisms of mTORC1 and mTORC2: Connected yet apart in cellular and molecular aspects. Adv Biol Regul 64:39–48
Kim M, Jo H, Kwon Y, Jeong MS, Jung HS, Kim Y, Jeoung D (2021) MiR-154-5p-MCP1 Axis Regulates Allergic Inflammation by Mediating Cellular Interactions. Front Immunol 12:2080
Li C, Han T, Li R, Fu L, Yue L (2020) miR-26a-5p mediates TLR signaling pathway by targeting CTGF in LPS-induced alveolar macrophage. Biosci Rep 40:BSR20192598
Liu Y, Zhang D-T, Liu X-G (2015) mTOR signaling in T cell immunity and autoimmunity. Int Rev Immunol 34:50–66
Lu QC, Rui ZH, Guo ZL, Xie W, Shan S, Ren T (2018) LncRNA-DANCR contributes to lung adenocarcinoma progression by sponging miR-496 to modulate mTOR expression. J Cell Mol Med 22:1527–1537
Mammana S, Bramanti P, Mazzon E, Cavalli E, Basile MS, Fagone P, Petralia MC, Mccubrey JA, Nicoletti F, Mangano K (2018) Preclinical evaluation of the PI3K/Akt/mTOR pathway in animal models of multiple sclerosis. Oncotarget 9:8263–8277
Meydan C, Madrer N, Soreq H (2020) The neat dance of COVID-19: NEAT1, DANCR, and co-modulated cholinergic RNAs link to inflammation. Front Immunol 11:590870
Polman CH, Reingold SC, Banwell B, Clanet M, Cohen JA, Filippi M, Fujihara K, Havrdova E, Hutchinson M, Kappos L (2011) Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol 69:292–302
Safa A, Arsang-Jang S, Taheri M, Omrani MD, Ghafouri-Fard S (2021) Dysregulation of NF-κB-Associated lncRNAs in multiple sclerosis patients. J Mol Neurosci 71:80–88
Sun I-H, Oh M-H, Zhao L, Patel CH, Arwood ML, Xu W, Tam AJ, Blosser RL, Wen J, Powell JD (2018) mTOR Complex 1 Signaling Regulates the Generation and Function of Central and Effector Foxp3(+) Regulatory T Cells. J Immunol (Baltimore, Md. : 1950) 201:481–492
Taherian-Esfahani Z, Taheri M, Dashti S, Kholghi-Oskooei V, Geranpayeh L, Ghafouri-Fard S (2019) Assessment of the expression pattern of mTOR-associated lncRNAs and their genomic variants in the patients with breast cancer. J Cell Physiol 234:22044–22056
Tong X, Gu P-C, Xu S-Z, Lin X-J (2015) Long non-coding RNA-DANCR in human circulating monocytes: a potential biomarker associated with postmenopausal osteoporosis. Biosci Biotechnol Biochem 79:732–737
Zhang S, Lin S, Tang Q, Yan Z (2021) Knockdown of miR-205-5p alleviates the inflammatory response in allergic rhinitis by targeting B-cell lymphoma 6. Mol Med Rep 24:1–11
Acknowledgements
This study was financially supported by Grant from Medical School of Shahid Beheshti University of Medical Sciences.
Author information
Authors and Affiliations
Contributions
SGF wrote the manuscript and revised it. MDO and MT supervised and designed the study. MG, MA, BMH and FE performed the experiment. SE analyzed the data. All authors read and approved the submitted version.
Corresponding authors
Ethics declarations
Ethics approval and consent to Participant
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent forms were obtained from all study participants. The study protocol was approved by the ethical committee of Shahid Beheshti University of Medical Sciences. All methods were performed in accordance with the relevant guidelines and regulations.
Consent of publication
Not applicable.
Competing Interest
The authors declare they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Akbari, M., Eshghyar, F., Gholipour, M. et al. Expression analysis of mTOR-associated lncRNAs in multiple sclerosis. Metab Brain Dis 37, 2061–2066 (2022). https://doi.org/10.1007/s11011-022-01010-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11011-022-01010-8