Abstract
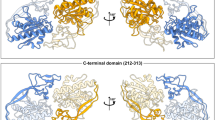
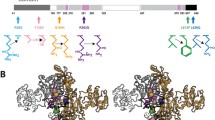
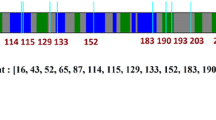
Aspartoacylase (ASPA) is a zinc-dependent abundant enzyme in the brain, which catalyzes the conversion of N-acetyl aspartate (NAA) into acetate and aspartate. Mutations in the ASPA gene are associated with the development of Canavan disease (CD), leading to the deficiency of ASPA activity. Patients with CD were characterized by degeneration of the white matter of the brain. We reported earlier on two patients with severe form of CD that both had two novel missense mutations in the ASPA: c.427 A > G; p. I143V and c.557 T > A; p. V186D (Zaki et al. 2017a), patient 1 harbored both mutations (p.I143V and p.V186D) in a heterozygous form together with four other mutations, and patient 2 had both mutations in homozygous form. Wijayasinghe et al. (2014) crystallized the 3D structures of four different ASPA mutants (p.K213E, p.Y231C, p.E285A, and p.F295S). In this study, we used in silico prediction methods and molecular dynamics simulation (MDS) to understand the structural impact of all these mutations. Moreover, we used molecular docking (MD) to investigate the binding patterns of the NAA substrate to the native and mutant proteins. Among the mutations, p.E285A (crystallized mutant) was predicted to be the most deleterious for the protein function and the least deleteriousness mutant was the p.I143V (novel mutant). Among the novel mutations, p.V186D was observed to be disruptive for both the zinc binding and NAA binding than the p.I143V. This study provides practical insights on the effect of these mutations on the ASPA function and might serve as a platform for drug design for CD treatment.








Similar content being viewed by others
Abbreviations
- ASPA:
-
Aspartoacylase
- CD:
-
Canavan disease
- 3D:
-
three-dimensional
- RMSD:
-
Root Mean Square Deviation
References
Ali SK, Sneha P, Priyadharshini Christy J et al (2016) Molecular dynamics-based analyses of the structural instability and secondary structure of the fibrinogen gamma chain protein with the D356V mutation. J Biomol Struct Dyn 35(12):2714–2724
Baslow MH, Guilfoyle DN (2013) Canavan disease, a rare early-onset human spongiform leukodystrophy: insights into its genesis and possible clinical interventions. Biochimie 95:946–956
Berendsen HJC, Postma JPM, van Gunsteren WF et al (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690
Birnbaum SM, Levintow L, Kingsley RB et al (1952) Specificity of amino acid acylases. J Biol Chem 194:455–470
Bitto E, Bingman CA, Wesenberg GE et al (2007) Structure of aspartoacylase, the brain enzyme impaired in Canavan disease. Proc Natl Acad Sci 104:456–461
Capriotti E, Fariselli P, Casadio R (2005) I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res 33:W306–W310
Capriotti E, Calabrese R, Casadio R (2006) Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics 22:2729–2734
Capriotti E, Altman RB, Bromberg Y (2013) Collective judgment predicts disease-associated single nucleotide variants. BMC Genomics 14(Suppl 3):S2
Cheatham TEI, Miller JL, Fox T et al (1995) Molecular dynamics simulations on solvated biomolecular systems: the particle mesh Ewald method leads to stable trajectories of DNA, RNA, and proteins. J Am Chem Soc 117:4193–4194
Chen C-W, Lin J, Chu Y-W (2013) iStable: off-the-shelf predictor integration for predicting protein stability changes. BMC Bioinf 14(Suppl 2):S5
Cheng J, Randall A, Baldi P (2006) Prediction of protein stability changes for single-site mutations using support vector machines. Proteins 62:1125–1132
Doss CGP, Alasmar DR, Bux RI et al (2016) Genetic epidemiology of glucose-6-phosphate dehydrogenase deficiency in the Arab world. Sci Rep 6:37284
George Priya Doss C, Chakraborty C, Narayan V, Thirumal Kumar D (2014) Computational approaches and resources in single amino acid substitutions analysis toward clinical research. Adv Protein Chem Struct Biol 94:365–423
Glaser F, Pupko T, Paz I et al (2003) ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics 19:163–164
Guex N, Peitsch, MC, (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 18(15):2714–2723
Hess B, Bekker H, Berendsen HJC, Fraaije JGEM (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18:1463–1472
Jakobs C, ten Brink HJ, Langelaar SA et al (1991) Stable isotope dilution analysis of N-acetylaspartic acid in CSF, blood, urine and amniotic fluid: accurate postnatal diagnosis and the potential for prenatal diagnosis of Canavan disease. J Inherit Metab Dis 14:653–660
Johnson AD, Handsaker RE, Pulit SL et al (2008) SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 24:2938–2939
Kaul R, Ping Gao G, Balamurugan K, Matalon R (1993) Cloning of the human Aspart-acylase cDNA and a common missense mutation in Canavan disease. Nat Genet 5:118–123
Kocak A, Yildiz M (2017) Docking, molecular dynamics and free energy studies on aspartoacylase mutations involved in Canavan disease. J Mol Graph Model 74:44–53
Kots ED, Khrenova MG, Lushchekina SV et al (2016) Modeling the complete catalytic cycle of Aspartoacylase. J PhysChem B 120:4221–4231
Kumar DT, Doss CGP (2016) Investigating the inhibitory effect of Wortmannin in the hotspot mutation at codon 1047 of PIK3CA kinase domain: a molecular docking and molecular dynamics approach. Adv Protein Chem Struct Biol 102:267–297
Le Coq J, Pavlovsky A, Malik R et al (2008) Examination of the mechanism of human brain Aspartoacylase through the binding of an intermediate. Analogue Biochemistry 47:3484–3492
Matalon R, Kaul R, Michals K (1993) Canavan disease: biochemical and molecular studies. J Inherit Metab Dis 16:744–752
Matalon R, Michals K, Kaul R (1995) Canavan disease: from spongy degeneration to molecular analysis. J Pediatr 127:511–517
Matalon R, Rady PL, Platt KA et al (2000) Knock-out mouse for Canavan disease: a model for gene transfer to the central nervous system. J Gene Med 2:165–175
Mi H, Thomas P (2009) PANTHER pathway: an ontology-based pathway database coupled with data analysis tools. Methods Mol Biol 563:123–140
Miyamoto S, Kollman PA (1992) Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J Comput Chem 13:952–962
Moore RA, Le Coq J, Faehnle CR, Viola RE (2003) Purification and preliminary characterization of brain aspartoacylase. Arch Biochem Biophys 413:1–8
Mosaeilhy A, Mohamed MM, George Priya Doss C et al (2017) Genotype-phenotype correlation in 18 Egyptian patients with glutaric acidemia type I. Metab Brain Dis. doi:https://doi.org/10.1007/s11011-017-0006-4
Ng PC, Henikoff S (2003) SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res 31:3812–3814
O’Boyle NM, Banck M, James CA et al (2011) Open babel: an open chemical toolbox. J Chem Inf 3:33
Parrinello M, Rahman A (1981) Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys 52:7182–7190
Peng Y, Norris J, Schwartz C, Alexov E (2016) Revealing the effects of missense mutations causing Snyder-Robinson syndrome on the stability and dimerization of Spermine synthase. Int J Mol Sci 17:E77
Pronk S, Páll S, Schulz R et al (2013) GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29:845–854
Salentin S, Schreiber S, Haupt VJ et al (2015) PLIP: fully automated protein-ligand interaction profiler. Nucleic Acids Res 43:W443–W447
Shastry BS (2007) SNPs in disease gene mapping, medicinal drug development and evolution. J Hum Genet 52:871–880
Sneha P, Doss CG (2016) Molecular dynamics: new frontier in personalized medicine. Adv Protein Chem Struct Biol 102:181–224
Sneha P, Kumar Thirumal D, Tanwar H et al (2017a) Structural analysis of G1691S variant in the human Filamin B gene responsible for Larsen syndrome: a comparative computational approach. J Cell Biochem 118:1900–1910
Sneha P, Thirumal Kumar D, George Priya Doss C et al (2017b) Determining the role of missense mutations in the POU domain of HNF1A that reduce the DNA-binding affinity: a computational approach. PLoS One 12(4):e0174953
Stefl S, Nishi H, Petukh M et al (2013) Molecular mechanisms of disease-causing missense mutations. J Mol Biol 425:3919–3936
Sujitha SP, Kumar DT, Doss CGP et al (2016) DNA repair gene (XRCC1) polymorphism (Arg399Gln) associated with schizophrenia in south Indian population: a genotypic and molecular dynamics study. PLoS One 11:e0147348
Surendran S, Michals-Matalon K, Quast MJ et al (2003) Canavan disease: a monogenic trait with complex genomic interaction. Mol Genet Metab 80:74–80
Thirumal Kumar D, George Priya Doss C (2016) Role of E542 and E545 missense mutations of PIK3CA in breast cancer: a comparative computational approach. J Biomol Struct Dyn 35:2745–2757
Thirumal Kumar D, Lavanya P, George Priya Doss C et al (2017) A molecular docking and dynamics approach to screen potent inhibitors against fosfomycin resistant enzyme in clinical Klebsiellapneumoniae. J Cell Biochem 45:777–787
Türkel N (2015) Stability constants of mixed ligand complexes of nickel(II) with adenine and some amino acids. Bioinorg Chem Appl 2015:374782
van Gunsteren WF, Billeter SR, Eising AA, Hünenberger PH, Krüger P, Mark AE, Scott WRP, Tironi IG (1996) Biomolecular simulation: the GROMOS96 manual and user guide. Vdf Hochschulverlag AG an der ETH Zürich, Zürich, pp 1–1042
Wang Z, Moult J (2001) SNPs, protein structure, and disease. Hum Mutat 17:263–270
Wang Z, Moult J (2003) Three-dimensional structural location and molecular functional effects of missense SNPs in the T cell receptor VB domain. Proteins Struct Funct Genet 53:748–757
Wijayasinghe YS, Pavlovsky AG, Viola RE (2014) Aspartoacylase catalytic deficiency as the cause of Canavan disease: a structural perspective. Biochemistry 53:4970–4978
Zaki OK, Krishnamoorthy N, El Abd HS et al (2017a) Two patients with Canavan disease and structural modeling of a novel mutation. Metab Brain Dis 32:171–177
Zaki OK, Priya Doss CG, Ali SA et al (2017b) Genotype–phenotype correlation in patients with isovaleric acidemia: comparative structural modelling and computational analysis of novel variants. Hum Mol Genet 12:e0174953
Zayed H (2015) Canavan disease: an Arab scenario. Gene. 560(1):9–14
Zhang Z, Norris J, Schwartz C et al (2011) In silico and in vitro investigations of the mutability of disease-causing missense mutation sites in Spermine synthase. PLoS One 6:e20373
Acknowledgments
Hatem Zayed is supported by the Qatar University grant QUUG-CAS-DHS-14/15-3.The authors also thank the VIT University management for their encouragement and provision of facilities.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interests
All authors declare no competing interests.
Electronic supplementary material
ESM 1
(DOCX 75.3 kb)
Rights and permissions
About this article
Cite this article
George Priya Doss, C., Zayed, H. Comparative computational assessment of the pathogenicity of mutations in the Aspartoacylase enzyme. Metab Brain Dis 32, 2105–2118 (2017). https://doi.org/10.1007/s11011-017-0090-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11011-017-0090-5