Abstract
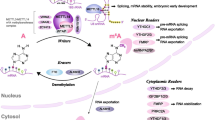
Epitranscriptomics, also known as “RNA epigenetics”, is a type of chemical modification that regulates RNA. RNA methylation is a significant discovery after DNA and histone methylation. The dynamic reversible process of m6A involves methyltransferases (writers), m6A binding proteins (readers), as well as demethylases (erasers). We summarized the current research status of m6A RNA methylation in the neural stem cells’ growth, synaptic and axonal function, brain development, learning and memory, neurodegenerative diseases, and glioblastoma. This review aims to provide a theoretical basis for studying the mechanism of m6A methylation and finding its potential therapeutic targets in nervous system diseases.



Similar content being viewed by others
Data availability
Not applicable.
Abbreviations
- ac4C:
-
N4-acetylcytidine
- AD:
-
Alzheimer's disease
- ADAR1:
-
Actadenosine deaminases acting on RNA-1
- ALKBH5:
-
AlkB homolog 5
- ALKBH9:
-
AlkB homolog 9
- aNSCs:
-
Adult neural stem cells
- CGCs:
-
Cerebellar granule cells
- DRG:
-
Adult dorsal root ganglion
- EGL:
-
External granular layer
- eIF3:
-
Eukaryotic translation initiation factor-3
- f6A:
-
N6-formyladenosine
- FMRP:
-
Fragment X mental retardation protein
- FTO:
-
Fat mass and obesity-associated protein
- GAK:
-
G-associated kinase
- GBM:
-
Glioblastoma
- GSCs:
-
Glioblastoma stem cells
- hm5C:
-
5-Hydroxymethylcytidine
- hm6A:
-
N6-hydroxymethyladenosine
- HNRNPA2B1:
-
Heterogeneous nuclear ribonucleoproteins A2/B1
- HNRNPC:
-
Heterogeneous ribonucleoprotein-C
- IAPs:
-
Intracisternal A-particles
- IGF2BPs:
-
Insulin-like growth factor 2 mRNA-binding proteins
- iPSCs:
-
Induced pluripotent stem cells
- L1:
-
Long interspersed element-1
- lncRNA:
-
Long non-coding RNA
- m1A:
-
N1-methyladenosine
- m5C:
-
5-Methylcytosine
- m6A:
-
N6-methyladenosine
- m6Am :
-
N6, 2'-O-dimethyladenosine
- MALAT1:
-
Metastasis-associated lung adenocarcinoma transcript 1
- METTL14:
-
Methyltransferase-like 14
- METTL16:
-
Methyltransferase-like protein 16
- METTL3:
-
Methyltransferase-like 3
- miRNA:
-
MicroRNA
- mRNA:
-
Messenger RNA
- NADP:
-
Nicotinamide adenine dinucleotide phosphate
- NMD:
-
Nonsense-mediated mRNA decay
- NMDA:
-
N-Methyl-D-aspartate
- NSCs:
-
Neural stem cells
- OPC:
-
Oligodendrocyte progenitor cells
- PD:
-
Parkinson's disease
- PolyA:
-
Polyadenosinic acid
- PRRC2A:
-
Proline-rich coiled-coil 2A
- RBM15:
-
RNA-binding motif protein 15
- rRNA:
-
Ribosomal RNA
- SAH:
-
S-Adenosyl homocysteine
- SAM:
-
S-Adenosylmethionine
- SENP1:
-
Sentrin/SUMO-specific protease 1
- SLC7A11:
-
Solute carrier 7A11
- SME:
-
Synaptic m6A epitranscriptome
- sncRNA:
-
Small non-coding RNA
- SNPC:
-
Substantia nigra pars compacta
- SNPs:
-
Single nucleotide polymorphisms
- SUMO1:
-
Small ubiquitin-like modifier 1
- SVZ:
-
Subventricular area
- TBI:
-
Traumatic brain injury
- tRNA:
-
Transfer RNA
- WTAP:
-
Wilms tumor 1-associating protein
- YTH:
-
YT521-B homology
- ZC3H13:
-
Zinc finger CCCH-type containing 13
- ZCCHC4:
-
CCHC zinc finger-containing protein
- Ψ :
-
Pseudouridine
References
Li X, Xiong X, Yi C (2016) Epitranscriptome sequencing technologies: decoding RNA modifications. Nat Methods 14:23–31. https://doi.org/10.1038/nmeth.4110
Zhao LY, Song J, Liu Y, Song CX, Yi C (2020) Mapping the epigenetic modifications of DNA and RNA. Protein Cell 11:792–808. https://doi.org/10.1007/s13238-020-00733-7
Boccaletto P, Machnicka MA, Purta E et al (2018) MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 46:D303-d307. https://doi.org/10.1093/nar/gkx1030
Nagarajan A, Janostiak R, Wajapeyee N (2019) Dot blot analysis for measuring global N6-methyladenosine modification of RNA. Methods Mol Biol 1870:263–271. https://doi.org/10.1007/978-1-4939-8808-2_20
Zhang C, Jia G (2018) Reversible RNA modification N1-methyladenosine (m1A) in mRNA and tRNA. Genom Proteom Bioinform 16:155–161. https://doi.org/10.1016/j.gpb.2018.03.003
Arango D, Sturgill D, Alhusaini N et al (2018) Acetylation of cytidine in mRNA promotes translation efficiency. Cell 175:1872-1886.e1824. https://doi.org/10.1016/j.cell.2018.10.030
Liu RJ, Long T, Li J, Li H, Wang ED (2017) Structural basis for substrate binding and catalytic mechanism of a human RNA:m5C methyltransferase NSun6. Nucleic Acids Res 45:6684–6697. https://doi.org/10.1093/nar/gkx473
Huber SM, van Delft P, Tanpure A, Miska EA, Balasubramanian S (2017) 2’-O-Methyl-5-hydroxymethylcytidine: a second oxidative derivative of 5-Methylcytidine in RNA. J Am Chem Soc 139:1766–1769. https://doi.org/10.1021/jacs.6b12180
Dominissini D, Moshitch-Moshkovitz S, Schwartz S et al (2012) Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485:201–206. https://doi.org/10.1038/nature11112
Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR (2012) Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149:1635–1646. https://doi.org/10.1016/j.cell.2012.05.003
Carlile TM, Martinez NM, Schaening C et al (2019) mRNA structure determines modification by pseudouridine synthase 1. Nat Chem Biol 15:966–974. https://doi.org/10.1038/s41589-019-0353-z
Chen XY, Zhang J, Zhu JS (2019) The role of m6A RNA methylation in human cancer. Mol Cancer 18:103. https://doi.org/10.1186/s12943-019-1033-z
Roundtree IA, Evans ME, Pan T, He C (2017) Dynamic RNA modifications in gene expression regulation. Cell 169:1187–1200. https://doi.org/10.1016/j.cell.2017.05.045
Brocard M, Ruggieri A, Locker N (2017) m6A RNA methylation, a new hallmark in virus-host interactions. J Gen Virol 98:2207–2214. https://doi.org/10.1099/jgv.0.000910
Roundtree IA, He C (2016) RNA epigenetics—chemical messages for posttranscriptional gene regulation. Curr Opin Chem Biol 30:46–51. https://doi.org/10.1016/j.cbpa.2015.10.024
Shi H, Wei J, He C (2019) Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell 74:640–650. https://doi.org/10.1016/j.molcel.2019.04.025
Jiang X, Liu B, Nie Z et al (2021) The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther 6:74. https://doi.org/10.1038/s41392-020-00450-x
Müller S, Glaß M, Singh AK et al (2019) IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A- and miRNA-dependent manner. Nucleic Acids Res 47:375–390. https://doi.org/10.1093/nar/gky1012
Tang C, Klukovich R, Peng H et al (2018) ALKBH5-dependent m6A demethylation controls splicing and stability of long 3’-UTR mRNAs in male germ cells. Proc Natl Acad Sci USA 115:E325-e333. https://doi.org/10.1073/pnas.1717794115
Fleming AM, Nguyen NLB, Burrows CJ (2019) Colocalization of m6A and G-quadruplex-forming sequences in viral RNA (HIV, Zika, Hepatitis B, and SV40) suggests topological control of adenosine N6-methylation. ACS Cent Sci 5:218–228. https://doi.org/10.1021/acscentsci.8b00963
Wu J, Frazier K, Zhang J, Gan Z, Wang T, Zhong X (2020) Emerging role of m6A RNA methylation in nutritional physiology and metabolism. Obes Rev 21:e12942. https://doi.org/10.1111/obr.12942
Dubey PK, Patil M, Singh S et al (2022) Increased m6A-RNA methylation and FTO suppression is associated with myocardial inflammation and dysfunction during endotoxemia in mice. Mol Cell Biochem 477:129–141. https://doi.org/10.1007/s11010-021-04267-2
Huang R, Zhang Y, Bai Y et al (2020) N6-methyladenosine modification of fatty acid amide hydrolase messenger RNA in circular RNA STAG1-regulated astrocyte dysfunction and depressive-like behaviors. Biol Psychiatry 88:392–404. https://doi.org/10.1016/j.biopsych.2020.02.018
Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Wu M (2019) Epigenetics in neurodevelopment: emerging role of circular RNA. Front Cell Neurosci 13:327. https://doi.org/10.3389/fncel.2019.00327
Hwang JY, Aromolaran KA, Zukin RS (2017) The emerging field of epigenetics in neurodegeneration and neuroprotection. Nat Rev Neurosci 18:347–361. https://doi.org/10.1038/nrn.2017.46
Nainar S, Marshall PR, Tyler CR, Spitale RC, Bredy TW (2016) Evolving insights into RNA modifications and their functional diversity in the brain. Nat Neurosci 19:1292–1298. https://doi.org/10.1038/nn.4378
Richard EM, Polla DL, Assir MZ et al (2019) Bi-allelic variants in METTL5 cause autosomal-recessive intellectual disability and microcephaly. Am J Hum Genet 105:869–878. https://doi.org/10.1016/j.ajhg.2019.09.007
Amort T, Rieder D, Wille A et al (2017) Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biol 18:1. https://doi.org/10.1186/s13059-016-1139-1
Vu LP, Pickering BF, Cheng Y et al (2017) The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med 23:1369–1376. https://doi.org/10.1038/nm.4416
Shi JB, Wang DY, Xia Q, Gao X (2020) The effects of m6A modification in central nervous system function and disease. Yi Chuan 42:1156–1167. https://doi.org/10.16288/j.yczz.20-233
Dermentzaki G, Lotti F (2020) New insights on the role of N6-methyladenosine RNA methylation in the physiology and pathology of the nervous system. Front Mol Biosci 7:555372. https://doi.org/10.3389/fmolb.2020.555372
Desrosiers R, Friderici K, Rottman F (1974) Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA 71:3971–3975. https://doi.org/10.1073/pnas.71.10.3971
Jia G, Fu Y, Zhao X et al (2011) N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 7:885–887. https://doi.org/10.1038/nchembio.687
Meyer KD, Jaffrey SR (2017) Rethinking m6A readers, writers, and erasers. Annu Rev Cell Dev Biol 33:319–342. https://doi.org/10.1146/annurev-cellbio-100616-060758
Du K, Zhang L, Lee T, Sun T (2019) m6A RNA methylation controls neural development and is involved in human diseases. Mol Neurobiol 56:1596–1606. https://doi.org/10.1007/s12035-018-1138-1
Liu J, Yue Y, Han D et al (2014) A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10:93–95. https://doi.org/10.1038/nchembio.1432
Lin S, Choe J, Du P, Triboulet R, Gregory RI (2016) The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell 62:335–345. https://doi.org/10.1016/j.molcel.2016.03.021
Wang P, Doxtader KA, Nam Y (2016) Structural basis for cooperative function of METTL3 and METTL14 methyltransferases. Mol Cell 63:306–317. https://doi.org/10.1016/j.molcel.2016.05.041
Wang X, Feng J, Xue Y et al (2016) Structural basis of N6-adenosine methylation by the METTL3 - METTL14 complex. Nature 534:575–578. https://doi.org/10.1038/nature18298
Chelmicki T, Roger E, Teissandier A et al (2021) m6A RNA methylation regulates the fate of endogenous retroviruses. Nature 591:312–316. https://doi.org/10.1038/s41586-020-03135-1
Du Y, Hou G, Zhang H et al (2018) SUMOylation of the m6A-RNA methyltransferase METTL3 modulates its function. Nucleic Acids Res 46:5195–5208. https://doi.org/10.1093/nar/gky156
Liu S, Zhuo L, Wang J et al (2020) METTL3 plays multiple functions in biological processes. Am J Cancer Res 10:1631–1646
Haussmann IU, Bodi Z, Sanchez-Moran E et al (2016) m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 540:301–304. https://doi.org/10.1038/nature20577
Patil DP, Chen CK, Pickering BF et al (2016) m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537:369–373. https://doi.org/10.1038/nature19342
Horiuchi K, Kawamura T, Iwanari H et al (2013) Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J Biol Chem 288:33292–33302. https://doi.org/10.1074/jbc.M113.500397
Pinto R, Vågbø CB, Jakobsson ME et al (2020) The human methyltransferase ZCCHC4 catalyses N6-methyladenosine modification of 28S ribosomal RNA. Nucl Acids Res 48:830–846. https://doi.org/10.1093/nar/gkz1147
Ruszkowska A, Ruszkowski M, Dauter Z, Brown JA (2018) Structural insights into the RNA methyltransferase domain of METTL16. Sci Rep 8:5311. https://doi.org/10.1038/s41598-018-23608-8
Qing Y, Dong L, Gao L et al (2021) R-2-hydroxyglutarate attenuates aerobic glycolysis in leukemia by targeting the FTO/m6A/PFKP/LDHB axis. Mol Cell. https://doi.org/10.1016/j.molcel.2020.12.026
Han Z, Wang X, Xu Z et al (2021) ALKBH5 regulates cardiomyocyte proliferation and heart regeneration by demethylating the mRNA of YTHDF1. Theranostics 11:3000–3016. https://doi.org/10.7150/thno.47354
Ontiveros RJ, Shen H, Stoute J et al (2020) Coordination of mRNA and tRNA methylations by TRMT10A. Proc Natl Acad Sci USA 117:7782–7791. https://doi.org/10.1073/pnas.1913448117
Zhang L, Wan Y, Zhang Z et al (2020) FTO demethylates m6A modifications in HOXB13 mRNA and promotes endometrial cancer metastasis by activating the WNT signalling pathway. RNA Biol 18:1265–1278. https://doi.org/10.1080/15476286.2020.1841458
Wang L, Song C, Wang N et al (2020) NADP modulates RNA m6A methylation and adipogenesis via enhancing FTO activity. Nat Chem Biol 16:1394–1402. https://doi.org/10.1038/s41589-020-0601-2
Hwang SY, Jung H, Mun S et al (2021) L1 retrotransposons exploit RNA m6A modification as an evolutionary driving force. Nat Commun 12:880. https://doi.org/10.1038/s41467-021-21197-1
Toh JDW, Crossley SWM, Bruemmer KJ et al (2020) Distinct RNAN-demethylation pathways catalyzed by nonheme iron ALKBH5 and FTO enzymes enable regulation of formaldehyde release rates. Proc Natl Acad Sci 117:25284–25292. https://doi.org/10.1073/pnas.2007349117
Liu J, Jia G (2014) Methylation modifications in eukaryotic messenger RNA. J Genet Genom 41:21–33. https://doi.org/10.1016/j.jgg.2013.10.002
Huff S, Tiwari SK, Gonzalez GM, Wang Y, Rana TM (2021) m6A-RNA demethylase FTO inhibitors impair self-renewal in glioblastoma stem cells. ACS Chem Biol 16:324–333. https://doi.org/10.1021/acschembio.0c00841
Tian S, Lai J, Yu T, Li Q, Chen Q (2020) Regulation of gene expression associated with the N6-methyladenosine (m6A) enzyme system and its significance in cancer. Front Oncol 10:623634. https://doi.org/10.3389/fonc.2020.623634
Wang X, Zhao Boxuan S, Roundtree Ian A et al (2015) N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161:1388–1399. https://doi.org/10.1016/j.cell.2015.05.014
Shi H, Wang X, Lu Z et al (2017) YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res 27:315–328. https://doi.org/10.1038/cr.2017.15
Zaccara S, Jaffrey SR (2020) A unified model for the function of YTHDF proteins in regulating m6A-modified mRNA. Cell 181:1582–1595. https://doi.org/10.1016/j.cell.2020.05.012
Roundtree IA, Luo GZ, Zhang Z et al (2017) YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife 6:e31311. https://doi.org/10.7554/eLife.31311
Zhou B, Liu C, Xu L et al (2020) N6-methyladenosine reader protein YT521-B homology domain-containing 2 suppresses liver steatosis by regulation of mRNA stability of lipogenic genes. Hepatology 73:91–103. https://doi.org/10.1002/hep.31220
Ma L, Chen T, Zhang X et al (2021) The m6A reader YTHDC2 inhibits lung adenocarcinoma tumorigenesis by suppressing SLC7A11-dependent antioxidant function. Redox Biol 38:101801. https://doi.org/10.1016/j.redox.2020.101801
Meyer KD, Patil DP, Zhou J et al (2015) 5’ UTR m6A promotes cap-independent translation. Cell 163:999–1010. https://doi.org/10.1016/j.cell.2015.10.012
Kratzat H, Mackens-Kiani T, Ameismeier M et al (2021) A structural inventory of native ribosomal ABCE1–43S pre-initiation complexes. Embo j 40:e105179. https://doi.org/10.15252/embj.2020105179
Alarcon CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF (2015) HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell 162:1299–1308. https://doi.org/10.1016/j.cell.2015.08.011
Wu B, Su S, Patil DP et al (2018) Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat Commun 9:420. https://doi.org/10.1038/s41467-017-02770-z
Huang H, Weng H, Sun W et al (2018) Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol 20:285–295. https://doi.org/10.1038/s41556-018-0045-z
Wu R, Li A, Sun B et al (2019) A novel m6A reader PRRC2A controls oligodendroglial specification and myelination. Cell Res 29:23–41. https://doi.org/10.1038/s41422-018-0113-8
Hsu PJ, Shi H, Zhu AC et al (2019) The RNA-binding protein FMRP facilitates the nuclear export of N6-methyladenosine-containing mRNAs. J Biol Chem 294:19889–19895. https://doi.org/10.1074/jbc.AC119.010078
Pendleton KE, Chen B, Liu K et al (2017) The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 169:824-835.e814. https://doi.org/10.1016/j.cell.2017.05.003
Bakhshinyan D, Savage N, Salim SK, Venugopal C, Singh SK (2020) The strange case of Jekyll and Hyde: parallels between neural stem cells and glioblastoma-initiating cells. Front Oncol 10:603738. https://doi.org/10.3389/fonc.2020.603738
Urbán N, Blomfield IM, Guillemot F (2019) Quiescence of adult mammalian neural stem cells: a highly regulated rest. Neuron 104:834–848. https://doi.org/10.1016/j.neuron.2019.09.026
Andreotti JP, Silva WN, Costa AC et al (2019) Neural stem cell niche heterogeneity. Semin Cell Dev Biol 95:42–53. https://doi.org/10.1016/j.semcdb.2019.01.005
Xing L, Cai Y, Yang T et al (2020) Epitranscriptomic m6A regulation following spinal cord injury. J Neurosci Res 99:843–857. https://doi.org/10.1002/jnr.24763
Chen J, Zhang YC, Huang C et al (2019) m6A regulates neurogenesis and neuronal development by modulating histone methyltransferase Ezh2. Genom Proteom Bioinform 17:154–168. https://doi.org/10.1016/j.gpb.2018.12.007
Wang Y, Li Y, Yue M et al (2018) N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat Neurosci 21:195–206. https://doi.org/10.1038/s41593-017-0057-1
Cao Y, Zhuang Y, Chen J et al (2020) Dynamic effects of FTO in regulating the proliferation and differentiation of adult neural stem cells of mice. Hum Mol Genet 29:727–735. https://doi.org/10.1093/hmg/ddz274
Heck AM, Russo J, Wilusz J, Nishimura EO, Wilusz CJ (2020) YTHDF2 destabilizes m6A-modified neural-specific RNAs to restrain differentiation in induced pluripotent stem cells. RNA 26:739–755. https://doi.org/10.1261/rna.073502.119
Stoeckli ET (2018) Understanding axon guidance: are we nearly there yet? Development 145:dev151415. https://doi.org/10.1242/dev.151415
von Kügelgen N, Chekulaeva M (2020) Conservation of a core neurite transcriptome across neuronal types and species. Wiley Interdiscip Rev RNA 11:e1590. https://doi.org/10.1002/wrna.1590
Kim SW, Kim KT (2020) Expression of genes involved in axon guidance: how much have we learned? Int J Mol Sci 21:3566. https://doi.org/10.3390/ijms21103566
Rizalar FS, Roosen DA, Haucke V (2021) A presynaptic perspective on transport and assembly mechanisms for synapse formation. Neuron 109:27–41. https://doi.org/10.1016/j.neuron.2020.09.038
Van Battum EY, Brignani S, Pasterkamp RJ (2015) Axon guidance proteins in neurological disorders. Lancet Neurol 14:532–546. https://doi.org/10.1016/s1474-4422(14)70257-1
Merkurjev D, Hong WT, Iida K et al (2018) Synaptic N6-methyladenosine (m6A) epitranscriptome reveals functional partitioning of localized transcripts. Nat Neurosci 21:1004–1014. https://doi.org/10.1038/s41593-018-0173-6
Koranda JL, Dore L, Shi H et al (2018) METTL14 is essential for epitranscriptomic regulation of striatal function and learning. Neuron 99:283–292. https://doi.org/10.1016/j.neuron.2018.06.007
Weng YL, Wang X, An R et al (2018) Epitranscriptomic m6A regulation of axon regeneration in the adult mammalian nervous system. Neuron 97:313–325. https://doi.org/10.1016/j.neuron.2017.12.036
Worpenberg L, Paolantoni C, Longhi S et al (2021) YTHDF is a N6-methyladenosine reader that modulates FMR1 target mRNA selection and restricts axonal growth in Drosophila. Embo j 40:e104975. https://doi.org/10.15252/embj.2020104975
Zhuang M, Li X, Zhu J et al (2019) The m6A reader YTHDF1 regulates axon guidance through translational control of Robo3.1 expression. Nucleic Acids Res 47:4765–4777. https://doi.org/10.1093/nar/gkz157
Yu J, Chen M, Huang H et al (2018) Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res 46:1412–1423. https://doi.org/10.1093/nar/gkx1182
Borsani E, Della Vedova AM, Rezzani R, Rodella LF, Cristini C (2019) Correlation between human nervous system development and acquisition of fetal skills: an overview. Brain Dev 41:225–233. https://doi.org/10.1016/j.braindev.2018.10.009
Vissers C, Sinha A, Ming GL, Song H (2020) The epitranscriptome in stem cell biology and neural development. Neurobiol Dis 146:105139. https://doi.org/10.1016/j.nbd.2020.105139
Shafik AM, Zhang F, Guo Z et al (2021) N6-methyladenosine dynamics in neurodevelopment and aging, and its potential role in Alzheimer’s disease. Genome Biol 22:17. https://doi.org/10.1186/s13059-020-02249-z
Yoon KJ, Ringeling FR, Vissers C et al (2017) Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell 171:877–889. https://doi.org/10.1016/j.cell.2017.09.003
Wang CX, Cui GS, Liu X et al (2018) METTL3-mediated m6A modification is required for cerebellar development. PLoS Biol 16:e2004880. https://doi.org/10.1371/journal.pbio.2004880
Li M, Zhao X, Wang W et al (2018) YTHDF2-mediated m6A mRNA clearance modulates neural development in mice. Genome Biol 19:69. https://doi.org/10.1186/s13059-018-1436-y
Li L, Zang L, Zhang F et al (2017) Fat mass and obesity-associated (FTO) protein regulates adult neurogenesis. Hum Mol Genet 26:2398–2411. https://doi.org/10.1093/hmg/ddx128
Du T, Li G, Yang J, Ma K (2020) RNA demethylase ALKBH5 is widely expressed in neurons and decreased during brain development. Brain Res Bull 163:150–159. https://doi.org/10.1016/j.brainresbull.2020.07.018
Van Damme S, De Fruyt N, Watteyne J et al (2021) Neuromodulatory pathways in learning and memory: lessons from invertebrates. J Neuroendocrinol 33:e12911. https://doi.org/10.1111/jne.12911
Ross TW, Easton A (2021) The hippocampal horizon: constructing and segmenting experience for episodic memory. Neurosci Biobehav Rev 132:181–196. https://doi.org/10.1016/j.neubiorev.2021.11.038
Rizzi-Wise CA, Wang DV (2021) Putting together pieces of the lateral septum: multifaceted functions and its neural pathways. eNeuro 8:315–321. https://doi.org/10.1523/eneuro.0315-21.2021
Todd TP, Fournier DI, Bucci DJ (2019) Retrosplenial cortex and its role in cue-specific learning and memory. Neurosci Biobehav Rev 107:713–728. https://doi.org/10.1016/j.neubiorev.2019.04.016
Baram TZ, Donato F, Holmes GL (2019) Construction and disruption of spatial memory networks during development. Learn Mem 26:206–218. https://doi.org/10.1101/lm.049239.118
Krüttner S, Caroni P (2018) m6A-epitranscriptome modulates memory strength. Cell Res 29:4–5. https://doi.org/10.1038/s41422-018-0121-8
Madugalle SU, Meyer K, Wang DO, Bredy TW (2020) RNA N6-methyladenosine and the regulation of RNA localization and function in the brain. Trends Neurosci 43:1011–1023. https://doi.org/10.1016/j.tins.2020.09.005
Engel M, Eggert C, Kaplick PM et al (2018) The role of m6A/m-RNA methylation in stress response regulation. Neuron 99:389–403. https://doi.org/10.1016/j.neuron.2018.07.009
Zhang Z, Wang M, Xie D et al (2018) METTL3-mediated N6-methyladenosine mRNA modification enhances long-term memory consolidation. Cell Res 28:1050–1061. https://doi.org/10.1038/s41422-018-0092-9
Zhang R, Zhang Y, Guo F et al (2022) Knockdown of METTL16 disrupts learning and memory by reducing the stability of MAT2A mRNA. Cell Death Discov 8:432. https://doi.org/10.1038/s41420-022-01220-0
Shi H, Zhang X, Weng YL et al (2018) m6A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature 563:249–253. https://doi.org/10.1038/s41586-018-0666-1
Kan L, Ott S, Joseph B et al (2021) A neural m6A/YTHDF pathway is required for learning and memory in Drosophila. Nat Commun 12:1458. https://doi.org/10.1038/s41467-021-21537-1
Yu J, Zhang Y, Ma H et al (2020) Epitranscriptomic profiling of N6-methyladenosine-related RNA methylation in rat cerebral cortex following traumatic brain injury. Mol Brain 13:11. https://doi.org/10.1186/s13041-020-0554-0
Ising C, Venegas C, Zhang S et al (2019) NLRP3 inflammasome activation drives tau pathology. Nature 575:669–673. https://doi.org/10.1038/s41586-019-1769-z
Scheltens P, Blennow K, Breteler MM et al (2016) Alzheimer’s disease. Lancet 388:505–517. https://doi.org/10.1016/s0140-6736(15)01124-1
Huang H, Camats-Perna J, Medeiros R, Anggono V, Widagdo J (2020) Altered expression of the m6A methyltransferase METTL3 in Alzheimer's disease. eNeuro 7: 0125–20. https://doi.org/10.1523/eneuro.0125-20.2020
Han M, Liu Z, Xu Y et al (2020) Abnormality of m6A mRNA methylation is involved in Alzheimer’s disease. Front Neurosci 14:98. https://doi.org/10.3389/fnins.2020.00098
Wei X, Cai M, Jin L (2020) The function of the metals in regulating epigenetics during Parkinson's disease. Front Genet 11:616083. https://doi.org/10.3389/fgene.2020.616083
Chen X, Yu C, Guo M et al (2019) Down-regulation of m6A mRNA methylation is involved in dopaminergic neuronal death. ACS Chem Neurosci 10:2355–2363. https://doi.org/10.1021/acschemneuro.8b00657
Qiu X, He H, Huang Y, Wang J, Xiao Y (2020) Genome-wide identification of m6A-associated single-nucleotide polymorphisms in Parkinson's disease. Neurosci Lett 737:135315. https://doi.org/10.1016/j.neulet.2020.135315
Yu Z, Huang L, Xia Y et al (2022) Analysis of m6A modification regulators in the substantia nigra and striatum of MPTP-induced Parkinson's disease mice. Neurosci Lett 791:136907. https://doi.org/10.1016/j.neulet.2022.136907
Le Rhun E, Preusser M, Roth P et al (2019) Molecular targeted therapy of glioblastoma. Cancer Treat Rev 80:101896. https://doi.org/10.1016/j.ctrv.2019.101896
Olivier C, Oliver L, Lalier L, Vallette FM (2020) Drug resistance in glioblastoma: the two faces of oxidative stress. Front Mol Biosci 7:620677. https://doi.org/10.3389/fmolb.2020.620677
Cui Q, Shi H, Ye P et al (2017) m6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep 18:2622–2634. https://doi.org/10.1016/j.celrep.2017.02.059
Tassinari V, Cesarini V, Tomaselli S et al (2021) ADAR1 is a new target of METTL3 and plays a pro-oncogenic role in glioblastoma by an editing-independent mechanism. Genome Biol 22:51. https://doi.org/10.1186/s13059-021-02271-9
Li F, Yi Y, Miao Y et al (2019) N6-methyladenosine modulates nonsense-mediated mrna decay in human glioblastoma. Cancer Res 79:5785–5798. https://doi.org/10.1158/0008-5472.Can-18-2868
Visvanathan A, Patil V, Arora A et al (2018) Essential role of METTL3-mediated m6A modification in glioma stem-like cells maintenance and radioresistance. Oncogene 37:522–533. https://doi.org/10.1038/onc.2017.351
Yarmishyn AA, Yang YP, Lu KH et al (2020) Musashi-1 promotes cancer stem cell properties of glioblastoma cells via upregulation of YTHDF1. Cancer Cell Int 20:597. https://doi.org/10.1186/s12935-020-01696-9
Fang R, Chen X, Zhang S et al (2021) EGFR/SRC/ERK-stabilized YTHDF2 promotes cholesterol dysregulation and invasive growth of glioblastoma. Nat Commun 12:177. https://doi.org/10.1038/s41467-020-20379-7
Wang LC, Chen SH, Shen XL et al (2020) M6A RNA methylation regulator hnrnpc contributes to tumorigenesis and predicts prognosis in glioblastoma multiforme. Front Oncol 10:536875. https://doi.org/10.3389/fonc.2020.536875
Deng X, Su R, Stanford S, Chen J (2018) Critical enzymatic functions of FTO in obesity and cancer. Front Endocrinol (Lausanne) 9:396. https://doi.org/10.3389/fendo.2018.00396
Kowalski-Chauvel A, Lacore MG, Arnauduc F et al (2020) The m6A RNA demethylase ALKBH5 promotes radioresistance and invasion capability of glioma stem cells. Cancers (Basel) 13:40. https://doi.org/10.3390/cancers13010040
Zhang C, Wang Y, Peng Y, Xu H, Zhou X (2020) METTL3 regulates inflammatory pain by modulating m6A-dependent pri-miR-365-3p processing. Faseb J 34:122–132. https://doi.org/10.1096/fj.201901555R
Kim H, Jang S (2021) RNA m6A methyltransferase METTL3 regulates spatial neural patterning in Xenopus. Mol Cell Biol 41:e0010421. https://doi.org/10.1128/mcb.00104-21
Wen L, Sun W, Xia D, Wang Y, Li J, Yang S (2020) The m6A methyltransferase METTL3 promotes LPS-induced microglia inflammation through TRAF6/NF-κB pathway. NeuroReport 33:243–251. https://doi.org/10.1097/wnr.0000000000001550
Xi Z, Xue Y, Zheng J, Liu X, Ma J, Liu Y (2016) WTAP expression predicts poor prognosis in malignant glioma patients. J Mol Neurosci 60:131–136. https://doi.org/10.1007/s12031-016-0788-6
Sun L, Ma L, Zhang H et al (2019) FTO deficiency reduces anxiety- and depression-like behaviors in mice via alterations in gut microbiota. Theranostics 9:721–733. https://doi.org/10.7150/thno.31562
Zhang L, Cheng Y, Xue Z et al (2021) Sevoflurane impairs m6A-mediated mRNA translation and leads to fine motor and cognitive deficits. Cell Biol Toxicol 38:347–369. https://doi.org/10.1007/s10565-021-09601-4
Zhang Z, Wang Q, Zhao X et al (2020) YTHDC1 mitigates ischemic stroke by promoting Akt phosphorylation through destabilizing PTEN mRNA. Cell Death Dis 11:977. https://doi.org/10.1038/s41419-020-03186-2
Kim HJ, Kim NC, Wang YD et al (2013) Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495:467–473. https://doi.org/10.1038/nature11922
Yin D, Kong C, Chen M (2020) Effect of hnRNPA2/B1 on the proliferation and apoptosis of glioma U251 cells via the regulation of AKT and STAT3 pathways. Biosci Rep 40:BSR20190318. https://doi.org/10.1042/bsr20190318
Deshaies JE, Shkreta L, Moszczynski AJ et al (2018) TDP-43 regulates the alternative splicing of hnRNP A1 to yield an aggregation-prone variant in amyotrophic lateral sclerosis. Brain 141:1320–1333. https://doi.org/10.1093/brain/awy062
Li ZW, Xue M, Zhu BX, Yue CL, Chen M, Qin HH (2019) microRNA-4500 inhibits human glioma cell progression by targeting IGF2BP1. Biochem Biophys Res Commun 513:800–806. https://doi.org/10.1016/j.bbrc.2019.04.058
Edens BM, Vissers C, Su J et al (2019) FMRP modulates neural differentiation through m6A-dependent mRNA nuclear export. Cell Rep 28:845–854. https://doi.org/10.1016/j.celrep.2019.06.072
Casingal CR, Kikkawa T, Inada H, Sasaki Y, Osumi N (2020) Identification of FMRP target mRNAs in the developmental brain: FMRP might coordinate Ras/MAPK, Wnt/β-catenin, and mTOR signaling during corticogenesis. Mol Brain 13:167. https://doi.org/10.1186/s13041-020-00706-1
Zhan X, Asmara H, Cheng N et al (2020) FMRP(1–297)-tat restores ion channel and synaptic function in a model of Fragile X syndrome. Nat Commun 11:2755. https://doi.org/10.1038/s41467-020-16250-4
Funding
This study was supported by the National Natural Science Foundation of China (81873351), and the Distinguished Young Scholars Project of Natural Science Foundation of Anhui Province in China (1908085J27).
Author information
Authors and Affiliations
Contributions
Nan Shao and Ting Ye analyzed the literature and drafted the manuscript. Weiting Xuan, Meng Zhang, Qian Chen and Liu Juan collected and screened the literature. Peng Zhou, Hang Song and Biao Cai conceived the study and revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shao, N., Ye, T., Xuan, W. et al. The effects of N6-methyladenosine RNA methylation on the nervous system. Mol Cell Biochem 478, 2657–2669 (2023). https://doi.org/10.1007/s11010-023-04691-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-023-04691-6