Abstract
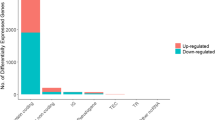
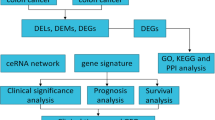
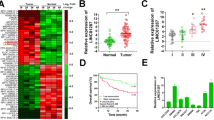
The purpose of this study was to identify the role of FEZF1-AS1 in colon cancer and predicted the underlying mechanism. We extracted sequencing data of colon cancer patients from The Cancer Genome Atlas database, identified the differential expression of long noncoding RNA, microRNA, and messenger RNA, constructed a competitive endogenous RNA network, and then analyzed prognosis. Then, we used the enrichment analysis databases for functional analysis. Finally, we studied the FEZF1-AS1/miR-92b-3p/ZIC5 axis. We detected the expression of FEZF1-AS1, miR-92b-3p, and ZIC5 via quantitative reverse transcription-PCR, transfected colon cancer cell RKO with lentivirus and conducted FEZF1-AS1 knockdown, and performed cancer-related functional assays. It indicated that many RNA in the competitive endogenous RNA network, such as ZIC5, were predicted to be related to overall survival of colon cancer patients, and enrichment analysis showed cancer-related signaling pathways, such as PI3K/AKT signaling pathway. The expression of FEZF1-AS1 and ZIC5 was significantly higher and that of miR-92b-3p was lower in the colon cancer than in the normal colon tissues. FEZF1-AS1 promoted the migration, proliferation, as well as invasion of RKO. According to the prediction, FEZF1-AS1 and ZIC5 might competitively bind to miR-92b-3p, leading to the weakening of the inhibitory impact of miR-92b-3p on ZIC5 and increasing expression of ZIC5, thus further activating the PI3K/AKT signaling pathway, which led to the occurrence and development of colon cancer. The study suggested that FEZF1-AS1 might be an effective diagnosis biomarker for colon cancer.










Similar content being viewed by others
Data availability
The datasets for this study can be found in the TCGA. Please see https://portal.gdc.cancer.gov/ for more details.
References
Siegel RL, Miller KD, Fuchs HE, Jemal A (2021) Cancer statistics, 2021. CA Cancer J Clin 71:7–33
Siegel RL, Miller KD, Goding Sauer A et al (2020) Colorectal cancer statistics, 2020. CA Cancer J Clin 70:145–164
Pan J, Cen L, Xu L et al (2020) Prevalence and risk factors for colorectal polyps in a Chinese population: a retrospective study. Sci Rep 10:6974
Wu H, Qin W, Lu S et al (2020) Long noncoding RNA ZFAS1 promoting small nucleolar RNA-mediated 2’-O-methylation via NOP58 recruitment in colorectal cancer. Mol Cancer 19:95
Wang H, Wang L, Xie Z et al (2020) Nitric oxide (NO) and NO synthases (NOS)-based targeted therapy for colon cancer. Cancers (Basel). 12:1881
Georgieva M, Gospodinova Z, Keremidarska-Markova M et al (2021) PEGylated nanographene oxide in combination with near-infrared laser irradiation as a smart nanocarrier in colon cancer targeted therapy. Pharmaceutics. 13:424
Du W, Yang X, He S et al (2021) Novel hyaluronic acid oligosaccharide-loaded and CD44v6-targeting oxaliplatin nanoparticles for the treatment of colorectal cancer. Drug Deliv 28:920–929
Lopes N, McIntyre C, Martin S et al (2021) Distinct metabolic programs established in the thymus control effector functions of γδ T cell subsets in tumor microenvironments. Nat Immunol 22:179–192
Wu X, Yang H, Chen X et al (2021) Nano-herb medicine and PDT induced synergistic immunotherapy for colon cancer treatment. Biomaterials 269:120654
Ding D, Zhong H, Liang R et al (2021) Multifunctional nanodrug mediates synergistic photodynamic therapy and MDSCs-targeting immunotherapy of colon cancer. Adv Sci (Weinh) 8:e2100712
Zhang X, Zhang H, Shen B, Sun XF (2019) Chromogranin-A expression as a novel biomarker for early diagnosis of colon cancer patients. Int J Mol Sci. 20:2919
Ludwig JA, Weinstein JN (2005) Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer 5:845–856
Weng M, Wu D, Yang C et al (2017) Noncoding RNAs in the development, diagnosis, and prognosis of colorectal cancer. Transl Res 181:108–120
Ezkurdia I, Juan D, Rodriguez JM et al (2014) Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes. Hum Mol Genet 23:5866–5878
Huarte M (2015) The emerging role of lncRNAs in cancer. Nat Med 21:1253–1261
Iyer MK, Niknafs YS, Malik R et al (2015) The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 47:199–208
Ponting CP, Oliver PL, Reik W (2009) Evolution and functions of long noncoding RNAs. Cell 136:629–641
Ning L, Li Z, Wei D et al (2017) LncRNA, NEAT1 is a prognosis biomarker and regulates cancer progression via epithelial-mesenchymal transition in clear cell renal cell carcinoma. Cancer Biomark 19:75–83
Xiao JN, Yan TH, Yu RM et al (2017) Long non-coding RNA UCA1 regulates the expression of Snail2 by miR-203 to promote hepatocellular carcinoma progression. J Cancer Res Clin Oncol 143:981
Zhang S, Wang Y, Chen M et al (2017) CXCL12 methylation-mediated epigenetic regulation of gene expression in papillary thyroid carcinoma. Sci Rep 7:44033
Li Y, Wu Z, Yuan J et al (2017) Long non-coding RNA MALAT1 promotes gastric cancer tumorigenicity and metastasis by regulating vasculogenic mimicry and angiogenesis. Cancer Lett 395:31–44
Bian Z, Zhang J, Li M et al (2018) LncRNA-FEZF1-AS1 promotes tumor proliferation and metastasis in colorectal cancer by regulating PKM2 signaling. Clin Cancer Res 24:4808–4819
Paul P, Chakraborty A, Sarkar D et al (2018) Interplay between miRNAs and human diseases. J Cell Physiol 233:2007–2018
Konovalova J, Gerasymchuk D, Parkkinen I et al (2019) Interplay between MicroRNAs and oxidative stress in neurodegenerative diseases. Int J Mol Sci. 20:6055
Chen C-Z (2005) MicroRNAs as oncogenes and tumor suppressors. N Engl J Med 353:1768–1771
Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6:857–866
Long M, Zhan M, Xu S et al (2017) miR-92b-3p acts as a tumor suppressor by targeting Gabra3 in pancreatic cancer. Mol Cancer 16:167
Tan X, Banerjee P, Liu X et al (2018) The epithelial-to-mesenchymal transition activator ZEB1 initiates a prometastatic competing endogenous RNA network. J Clin Invest 128:1267–1282
Chang L, Yuan Y, Li C et al (2016) Upregulation of SNHG6 regulates ZEB1 expression by competitively binding miR-101-3p and interacting with UPF1 in hepatocellular carcinoma. Cancer Lett 383:183–194
Zhan W, Liao X, Chen Z et al (2020) LINC00858 promotes colorectal cancer by sponging miR-4766-5p to regulate PAK2. Cell Biol Toxicol 36:333–347
Zhuang M, Zhao S, Jiang Z et al (2019) MALAT1 sponges miR-106b-5p to promote the invasion and metastasis of colorectal cancer via SLAIN2 enhanced microtubules mobility. EBioMedicine 41:286–298
Liang H, Yu T, Han Y et al (2018) LncRNA PTAR promotes EMT and invasion-metastasis in serous ovarian cancer by competitively binding miR-101-3p to regulate ZEB1 expression. Mol Cancer 17:119
Li C, Wan L, Liu Z et al (2018) Long non-coding RNA XIST promotes TGF-β-induced epithelial-mesenchymal transition by regulating miR-367/141-ZEB2 axis in non-small-cell lung cancer. Cancer Lett 418:185–195
Weinstein JN, Collisson EA, Mills GB et al (2013) The cancer genome atlas pan-cancer analysis project. Nat Genet 45:1113–1120
Tippmann S (2015) Programming tools: adventures with R. Nature 517:109–110
Ritchie ME, Phipson B, Wu D et al (2015) Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47
Lin Y, Liu T, Cui T et al (2020) RNAInter in 2020: RNA interactome repository with increased coverage and annotation. Nucleic Acids Res 48:D189–D197
Shannon P, Markiel A, Ozier O et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504
Yu G, Wang L-G, Han Y, He Q-Y (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16:284–287
Subramanian A, Tamayo P, Mootha VK et al (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–15550
Zhou R-S, Zhang E-X, Sun Q-F et al (2019) Integrated analysis of lncRNA-miRNA-mRNA ceRNA network in squamous cell carcinoma of tongue. BMC Cancer 19:779
Wang CC, Han CD, Zhao Q, Chen X (2021) Circular RNAs and complex diseases: from experimental results to computational models. Brief Bioinform. https://doi.org/10.1093/bib/bbab286
Bhan A, Soleimani M, Mandal SS (2017) Long noncoding RNA and cancer: a new paradigm. Cancer Res 77:3965–3981
Murugan AK, Munirajan AK, Alzahrani AS (2018) Long noncoding RNAs: emerging players in thyroid cancer pathogenesis. Endocr Relat Cancer 25:R59–R82
Yao J, Yang Z, Yang J et al (2021) Long non-coding RNA FEZF1-AS1 promotes the proliferation and metastasis of hepatocellular carcinoma via targeting miR-107/Wnt/β-catenin axis. Aging (Albany NY). 13:13726
Dasgupta P, Kulkarni P, Majid S et al (2020) LncRNA CDKN2B-AS1/miR-141/cyclin D network regulates tumor progression and metastasis of renal cell carcinoma. Cell Death Dis 11:660
Fang J, Ding Z (2020) SNAI1 is a prognostic biomarker and correlated with immune infiltrates in gastrointestinal cancers. Aging (Albany NY) 12:17167–17208
Satow R, Inagaki S, Kato C et al (2017) Identification of zinc finger protein of the cerebellum 5 as a survival factor of prostate and colorectal cancer cells. Cancer Sci 108:2405–2412
Dang W, Zhu Z (2020) MicroRNA-1249 targets four-jointed box kinase 1 and reduces cell proliferation, migration and invasion of colon adenocarcinoma. J Gene Med 22:e3183
Lin K, Huang J, Luo H et al (2020) Development of a prognostic index and screening of potential biomarkers based on immunogenomic landscape analysis of colorectal cancer. Aging (Albany NY) 12:5832–5857
The Gene Ontology Consortium (2015) Gene ontology consortium: going forward. Nucleic Acids Res 43:D1049–D1056
Chen L, Zhang YH, Wang S et al (2017) Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PLoS ONE 12:e0184129
Kanehisa M, Furumichi M, Tanabe M et al (2017) KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45:D353–D361
Abdelwahed KS, Siddique AB, Mohyeldin MM et al (2020) Pseurotin A as a novel suppressor of hormone dependent breast cancer progression and recurrence by inhibiting PCSK9 secretion and interaction with LDL receptor. Pharmacol Res 158:104847
Canuel M, Sun X, Asselin M-C et al (2013) Proprotein convertase subtilisin/kexin type 9 (PCSK9) can mediate degradation of the low density lipoprotein receptor-related protein 1 (LRP-1). PLoS ONE 8:e64145
Godthelp BC, Wiegant WW, van Duijn-Goedhart A et al (2002) Mammalian Rad51C contributes to DNA cross-link resistance, sister chromatid cohesion and genomic stability. Nucleic Acids Res 30:2172–2182
Connell PP, Siddiqui N, Hoffman S et al (2004) A hot spot for RAD51C interactions revealed by a peptide that sensitizes cells to cisplatin. Cancer Res 64:3002–3005
Wiese C, Collins DW, Albala JS et al (2002) Interactions involving the Rad51 paralogs Rad51C and XRCC3 in human cells. Nucleic Acids Res 30:1001–1008
Liu N, Schild D, Thelen MP, Thompson LH (2002) Involvement of Rad51C in two distinct protein complexes of Rad51 paralogs in human cells. Nucleic Acids Res 30:1009–1015
Böhme I, Beck-Sickinger AG (2009) Illuminating the life of GPCRs. Cell Commun Signal 7:16
Vogelstein B, Papadopoulos N, Velculescu VE et al (2013) Cancer genome landscapes. Science 339:1546–1558
Baldwin AS (1996) The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu Rev Immunol 14:649–683
Li F, Zhang J, Arfuso F et al (2015) NF-κB in cancer therapy. Arch Toxicol 89:711–731
Kojima M, Morisaki T, Sasaki N et al (2004) Increased nuclear factor-kB activation in human colorectal carcinoma and its correlation with tumor progression. Anticancer Res 24:675–681
Liu B, Xu T, Xu X et al (2018) Biglycan promotes the chemotherapy resistance of colon cancer by activating NF-κB signal transduction. Mol Cell Biochem 449:285–294
Cross MJ, Dixelius J, Matsumoto T, Claesson-Welsh L (2003) VEGF-receptor signal transduction. Trends Biochem Sci 28:488–494
Fruman DA, Chiu H, Hopkins BD et al (2017) The PI3K pathway in human disease. Cell 170:605–635
Yang Q, Jiang W, Hou P (2019) Emerging role of PI3K/AKT in tumor-related epigenetic regulation. Semin Cancer Biol 59:112–124
Bishnupuri KS, Alvarado DM, Khouri AN et al (2019) IDO1 and kynurenine pathway metabolites activate PI3K-Akt signaling in the neoplastic colon epithelium to promote cancer cell proliferation and inhibit apoptosis. Cancer Res 79:1138–1150
Johnson L, Mercer K, Greenbaum D et al (2001) Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature 410:1111–1116
Zimmermann G, Papke B, Ismail S et al (2013) Small molecule inhibition of the KRAS-PDEδ interaction impairs oncogenic KRAS signalling. Nature 497:638–642
Li JH, Liu S, Zhou H et al (2014) starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic acids Res 42:D92–D97
Liu YW, Xia R, Lu K et al (2017) LincRNAFEZF1-AS1 represses p21 expression to promote gastric cancer proliferation through LSD1-Mediated H3K4me2 demethylation. Mol Cancer 16:39
Chen N, Guo D, Xu Q et al (2016) Long non-coding RNA FEZF1-AS1 facilitates cell proliferation and migration in colorectal carcinoma. Oncotarget 7:11271–11283
Zhou Y, Xu S, Xia H et al (2019) Long noncoding RNA FEZF1-AS1 in human cancers. Clin Chim Acta 497:20–26
Zhang T, Yu S, Zhao S (2021) LncRNA FEZF1-AS1 promotes colorectal cancer progression through regulating the miR-363-3p/PRRX1 pathway. Adv Clin Exp Med 30:839–848
Acknowledgements
We express our sincere gratitude to the TCGA database, RNAInter database, Starbase for data acquisition, and Cuiying Biomedical Research Center for their support of our laboratory equipment and laboratory technology.
Funding
The study was supported by the National Natural Science Foundation of China [grant number 81770525]; Cuiying Scientific and Technological Innovation Program of Lanzhou University Second Hospital [grant number CY2021-QN-A05]; Cuiying Scientific and Technological Key Cultivation Program of Lanzhou University Second Hospital [grant number CY2018-ZD01]; Lanzhou Science and technology project [grant number 2022-ZD-104]; Science and technology project of Gansu Province [grant number 21JR1RA148]; Natural Science Foundation of Gansu Province [22JR5RA969]; and Industrial support and guidance project for institutions of higher learning in Gansu province (2019C-21).
Author information
Authors and Affiliations
Contributions
The original study design was undertaken by DZ, XY, and PW. Data were analyzed by XY and XM. Experiments were performed by XY, XS, ZW, and ZW. The results were analyzed by FW and XY.
Corresponding author
Ethics declarations
Competing interest
The authors declare that there are no competing interests.
Ethical approval
Ethical approval to the human study was provided by Lanzhou University Second Hospital medical ethics committee (APPROVAL NUMBER 2021A-468).
Informed consent
We obtained the informed consents from the colon cancer patients.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, X., Wu, P., Wang, Z. et al. Constructed the ceRNA network and predicted a FEZF1-AS1/miR-92b-3p/ZIC5 axis in colon cancer. Mol Cell Biochem 478, 1083–1097 (2023). https://doi.org/10.1007/s11010-022-04578-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-022-04578-y