Abstract
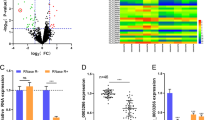
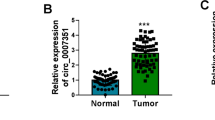
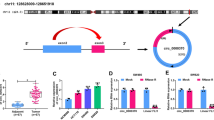
The purpose of this paper was to explore the role of circ_0056618 and associated mechanisms in colorectal cancer (CRC). The expression of circ_0056618, proline rich and Gla domain 4 (PRRG4) mRNA and miR-411-5p was measured by quantitative real-time PCR (qPCR).The protein levels of PRRG4 and epithelial-mesenchymal transition (EMT)-related markers were detected by western blot. Cell proliferation was assessed by cell counting kit-8, EdU, and colony formation assays. Cell migration and invasion were assessed by transwell assay. Cell apoptosis was detected by flow cytometry assay. The putative relationship between miR-411-5p and circ_0056618 or PRRG4 was verified by dual-luciferase reporter assay. The effects of circ_0056618 on tumor growth in vivo were determined by animal study. Circ_0056618 and PRRG4 was upregulated, while miR-411-5p was downregulated in CRC tumor tissues and cells. Circ_0056618 knockdown or PRRG4 knockdown inhibited CRC cell proliferation, migration/invasion, EMT, and survival. Circ_0056618 positively modulated PRRG4 expression by targeting miR-411-5p. MiR-411-5p absence or PRRG4 overexpression could rescue circ_0056618 knockdown-induced inhibition on proliferation, migration/invasion, and EMT in CRC cells. Animal assay showed circ_0056618 knockdown impeded tumor growth in vivo. Circ_0056618 promoted CRC growth and development by upregulating PRRG4 expression via competitively targeting miR-411-5p.








Similar content being viewed by others
Data availability
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
References
Sung H, Ferlay J, Siegel RL et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA 71:209–249
Lizarbe MA, Calle-Espinosa J, Fernandez-Lizarbe E et al (2017) Colorectal cancer: from the genetic model to posttranscriptional regulation by noncoding RNAs. BioMed Res Int. https://doi.org/10.1155/2017/7354260
Kuipers EJ, Grady WM, Lieberman D, et al (2015) Colorectal cancer. Nat Rev Dis Primers 1: 15065.
Lech G, Slotwinski R, Slodkowski M, Krasnodebski IW (2016) Colorectal cancer tumour markers and biomarkers: Recent therapeutic advances. World J Gastroenterol 22:1745–1755
Chen B, Huang S (2018) Circular RNA: An emerging non-coding RNA as a regulator and biomarker in cancer. Cancer Lett 418:41–50
Suzuki H, Tsukahara T (2014) A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci 15:9331–9342
Zhao ZJ, Shen J (2017) Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol 14:514–521
Yang H, Li X, Meng Q et al (2020) CircPTK2 (hsa_circ_0005273) as a novel therapeutic target for metastatic colorectal cancer. Mol Cancer 19:13
Chen LY, Wang L, Ren YX et al (2020) The circular RNA circ-ERBIN promotes growth and metastasis of colorectal cancer by miR-125a-5p and miR-138-5p/4EBP-1 mediated cap-independent HIF-1alpha translation. Mol Cancer 19:164
Zheng X, Chen L, Zhou Y et al (2019) A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer 18:47
Zheng X, Ma YF, Zhang XR, Li Y, Zhao HH, Han SG (2020) Circ_0056618 promoted cell proliferation, migration and angiogenesis through sponging with miR-206 and upregulating CXCR4 and VEGF-A in colorectal cancer. Eur Rev Med Pharmacol Sci 24:4190–4202
Harrandah AM, Mora RA, Chan EKL (2018) Emerging microRNAs in cancer diagnosis, progression, and immune surveillance. Cancer Lett 438:126–132
Tang Q, Hann SS (2020) Biological roles and mechanisms of circular RNA in human cancers. Onco Targets Ther 13:2067–2092
Chipman LB, Pasquinelli AE (2019) miRNA targeting: growing beyond the seed. Trends Genet 35:215–222
Zhao J, Xu J, Zhang R (2018) MicroRNA-411 inhibits malignant biological behaviours of colorectal cancer cells by directly targeting PIK3R3. Oncol Rep 39:633–642
Marshall KW, Mohr S, Khettabi FE et al (2010) A blood-based biomarker panel for stratifying current risk for colorectal cancer. Int J Cancer 126:1177–1186
Yip KT, Das PK, Suria D, Lim CR, Ng GH, Liew CC (2010) A case-controlled validation study of a blood-based seven-gene biomarker panel for colorectal cancer in Malaysia. J exp clinic cancer res 29:128
Ke B, Ye K, Cheng S (2020) ALKBH2 inhibition alleviates malignancy in colorectal cancer by regulating BMI1-mediated activation of NF-kappaB pathway. World J Surg Oncol 18:328
Feng S, Luo S, Ji C, Shi J (2020) miR-29c-3p regulates proliferation and migration in ovarian cancer by targeting KIF4A. World J Surg Oncol 18:315
Li H, Yao G, Feng B, Lu X, Fan Y (2018) Circ_0056618 and CXCR4 act as competing endogenous in gastric cancer by regulating miR-206. J Cell Biochem 119:9543–9551
Li S, Zhang L, Li S, Zhao H, Chen Y (2021) Curcumin suppresses the progression of gastric cancer by regulating circ_0056618/miR-194-5p axis. Open Life Sci 16:937–949
Li L, Li W (2015) Epithelial-mesenchymal transition in human cancer: comprehensive reprogramming of metabolism, epigenetics, and differentiation. Pharmacol Ther 150:33–46
Greaves D, Calle Y (2022) Epithelial Mesenchymal Transition (EMT) and associated invasive adhesions in solid and haematological tumours. Cells 11(4):649
Zhang Y, Xu G, Liu G et al (2016) miR-411-5p inhibits proliferation and metastasis of breast cancer cell via targeting GRB2. Biochem Biophys Res Commun 476:607–613
Xia LH, Yan QH, Sun QD, Gao YP (2018) MiR-411-5p acts as a tumor suppressor in non-small cell lung cancer through targeting PUM1. Eur Rev Med Pharmacol Sci 22:5546–5553
He F, Zu D, Lan C, Niu J, Nie X (2020) hsa-microRNA-411-5p regulates proliferation, migration and invasion by targeting the hyaluronan mediated motility receptor in ovarian cancer. Exp Ther Med 20:1899–1906
Zhang L, Qin Y, Wu G et al (2020) PRRG4 promotes breast cancer metastasis through the recruitment of NEDD4 and downregulation of Robo1. Oncogene 39(49):7196–7208
Funding
No funding was received.
Author information
Authors and Affiliations
Contributions
WC and YL: conceptualization and methodology; YZ, CL and BS: formal analysis and data curation; BZ and WC: validation and investigation; BZ, WC and YL: writing—original draft preparation and writing—review and editing; all authors: approval of final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval
The present study was approved by the ethical review committee of North China University of Science and Technology Affiliated Hospital. Written informed consent was obtained from all enrolled patients.
Consent for publication
The results presented in this paper have not been published preciously in whole or in part.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11010_2022_4525_MOESM1_ESM.tif
Supplementary file1 (TIF 44868 KB) Fig. S1 The representative images of colony formation, cell migration, invasion, and apoptosis in SW620 and HCT116 cells with si-NC, si-circ_0056618, si-circ_0056618+anti-NC, or si-circ_0056618+anti-miR-411-5p transfection.
11010_2022_4525_MOESM2_ESM.tif
Supplementary file2 (TIF 46504 KB) Fig. S2 The representative images of colony formation, cell migration, invasion, and apoptosis in SW620 and HCT116 cells with si-NC, si-circ_0056618, si-circ_0056618+pcDNA, or si-circ_0056618+pcDNA-PRRG4 transfection.
Rights and permissions
About this article
Cite this article
Zhang, B., Cao, W., Liu, Y. et al. Circ_0056618 enhances PRRG4 expression by competitively binding to miR-411-5p to promote the malignant progression of colorectal cancer. Mol Cell Biochem 478, 503–516 (2023). https://doi.org/10.1007/s11010-022-04525-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-022-04525-x