Abstract
Background
Circular RNAs (circRNAs) feature prominently in tumor progression. However, the biological function and molecular mechanism of circ_0003266 in colorectal cancer (CRC) require further investigation.
Methods
Circ_0003266 expression in 46 pairs CRC tissues / adjacent tissues, and CRC cell lines was detected by quantitative real-time polymerase chain reaction (qRT-PCR); after circ_0003266 was overexpressed or knocked down in CRC cells, cell proliferation, apoptosis, migration, and invasion were evaluated by the cell counting kit-8 (CCK-8), flow cytometry, and Transwell assays, respectively; the interaction among circ_0003266, miR-503-5p, and programmed cell death 4 (PDCD4) was confirmed using bioinformatics analysis and dual-luciferase reporter assay; PDCD4 protein expression in CRC cells was quantified using Western blot.
Results
Circ_0003266 was significantly lowly expressed in CRC tissues and cell lines. Circ_0003266 overexpression markedly repressed CRC cell proliferation, migration, and invasion, and accelerated the cell apoptosis, but its overexpression promoted the malignant phenotypes of CRC cells. PDCD4 was a direct target of miR-503-5p and circ_0003266 promoted PDCD4 expression by competitively sponging miR-503-5p.
Conclusion
Circ_0003266 suppresses the CRC progression via sponging miR-503-5p and regulating PDCD4 expressions, which suggests that circ_0003266 may serve as a novel target for the treatment of CRC.
Similar content being viewed by others
Background
Colorectal cancer (CRC) is a common cancer, and its mortality ranks among cancers worldwide, with nearly 900,000 deaths each year [1]. Although great progress has been made in the diagnosis and treatment in recent years, the therapeutic effect of some patients with CRC is not good due to high frequency of metastasis and recurrence [2]. Besides, the incidence of CRC is increasing among people under 45 years old [3]. The research on circular RNAs (circRNAs), microRNA (miRNA), and their regulatory mechanism in CRC may provide novel diagnostic biomarkers and therapeutic targets for CRC, which may help improve the prognosis of the patients with CRC [4, 5].
CircRNA is a class of non-coding RNA derived from the reverse splicing of the precursor mRNA, which is with a covalent closed-loop structure formed by splicing the 5′ end of one exon with the 3′ end of another exon [6]. Many circRNAs are dysregulated in diverse diseases [7, 8]. For example, hsa_circ_0013958 expression is up-regulated in lung adenocarcinoma tissues, cells, and plasma of the patients, which is positively correlated with TNM stage and lymphatic metastasis; functionally, circ_0013958 accelerates the proliferation and invasion of lung adenocarcinoma cells and inhibits the apoptosis [9]. Some circRNAs are abnormally expressed in CRC tissues, in which circRNAs with down-regulated expression levels, such as circ_0008287, circ_0069865, and circVPS13–1, are tumor suppressors, while circRNAs with up-regulated expression levels, such as circMGAT5, circ_0000724, and circAATF-1, act as tumor promoters [10]. However, the function of circ_0003266 in CRC awaits further study.
MicroRNAs (miRNAs), an endogenous RNA with about 20–22 nt, is involved in regulating many physiological and pathological processes. CircRNAs can exert their function via sponging miRNA, competitively combine with the corresponding miRNA through base pairing, and regulate gene expression at the post-transcriptional level [11]. For example, circAPLP2 activates Notch signaling pathway in CRC by targeting miR-101-3p, thus promoting tumor proliferation and metastasis [12]. CircAGFG1 drives the metastasis of CRC by modulating the YY1/CTNNB1 axis via sponging miR-4262 and miR-185-5p [13]. Reportedly, miR-503 promotes the migration and invasion of CRC cells by regulating programmed cell death 4 (PDCD4) [14]. However, whether miR-503/PDCD4 axis is involved in a competitively endogenous RNA (ceRNA) network in CRC is still obscure.
In this work, we used circRNA microarray to identify the abnormal expression of circRNAs in CRC tissues. We demonstrated that, circ_0003266 expression was significantly down-regulated in CRC. Functionally, circ_0003266 impeded the proliferation and metastasis of CRC cells and promoted apoptosis by regulating miR-503-5p/PDCD4 pathway.
Methods
Tissue samples
The study enrolled 46 CRC patients (22 males and 24 females, aged from 23 to 60 years) recruited between 2018 and 2019 from the Yichang Central People’s Hospital. All CRC patients who had undergone surgery without chemotherapy or radiotherapy were diagnosed by pathological examination. The cancerous and paracancerous tissues (more than 2 cm from the edge of the tumor) were collected and immediately stored in liquid nitrogen. Tumor histological grading and staging were performed according to the World Health Organization classification criteria and the Tumor Node Metastasis system. This study was endorsed by the Institutional Ethics Committee of Yichang Central People’s Hospital, and written informed consents were obtained from all patients before the research.
Expression profile analysis of circRNAs
CircRNA expression profile data were downloaded from Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). Using keywords (“circRNA” and “colorectal cancer”) in the GEO database, we searched the circRNAs microarray related with CRC and found the dataset GSE142837. We used GEO2R online analysis tool to get log fold change and adjusted P-value. Excel was used to screen out the circRNAs with P < 0.05 and |log2fold change (FC)| > 1 in CRC tissues (v.s. non-tumor tissues).
Cell culture
Human normal colonic epithelial cells (NCM460) and CRC cell lines (HT29, SW480, HCT-116, Lovo, and DLD-1) were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All CRC cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS, Gibco, Carlsbad, CA, USA), 100 U/ml penicillin and 100 μg/ml streptomycin (Sigma, St. Louis, MO, USA) at 37 °C in 5% CO2.
Cell transfection
The overexpression vectors of circ_0003266 and PDCD4 were constructed using pcDNA3.1 vector. siRNA targeting circ_0003266 (si-circ_0003266), miR-503-5p mimic and inhibitor, and their corresponding controls were purchased from GenePharma (Shanghai, China). The negative control (si-NC or miR-control) was adopted as the control vectors. The above mentioned oligonucleotides or plasmids (50 nM) and Lipofectamine™ 2000 reagents (Invitrogen, Carlsbad, CA, USA) were diluted using 100 μL of Opti-MEM medium (Invitrogen, Carlsbad, CA, USA), respectively, and incubated for 2 min at room temperature. Then they were mixed and incubated at room temperature for 20 min. The mixture was then added to a 6-well plate (containing 3 × 105 cells/well). 48 h after the transfection, the transfection efficiency was detected.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated using TRIzol (Vazyme, Nanjing, China). cDNA synthesis was conducted using the TaqMan MicroRNA reverse transcription kit (Applied Biosystems, Foster City, CA) for miR-503-5p and PrimeScript RT Master Mix Kit (Takara Biotechnology Co., Ltd., Dalian, China) were used for preparing the cDNA to detect PDCD4 and circ_0003266. Then quantitative PCR was performed, and circ_0003266 and PDCD4 expression levels were determined by SYBR SYBR Premix Ex Taq II (Takara, Dalian, China), and miR-503-5p expression was quantified by stem-loop primer SYBR Green qRT-PCR (Synbio Tech, Suzhou, China). GAPDH and U6 worked as internal controls for circRNA/mRNA and miRNA, respectively. qRT-PCR was operated on an ABI 7500 Fast Real-Time PCR System (Applied Biosystems, Waltham, MA, UK), and relative expression levels were calculated by 2−ΔΔCT method. Primer sequences are listed in Table 1.
RNase R resistance analysis of circRNAs
To confirm the circular property of circ_0003266, 2 μg of total RNAs was treated with or without 3 U/mg RNase R (Epicentre Technologies, Madison, WI, USA) for 30 min at 37 °C in RNase R reaction buffer. Then the expression level of circ_0003266 was detected by qRT-PCR.
Western blotting
Total protein was extracted by RIPA lysis buffer (Beyotime, Shanghai, China). Subsequently, protein concentration was measured using a bicinchoninic assay. Then 30 μg of protein / lane was separated via 10% SDS-PAGE and then transferred onto a PVDF membrane (Millipore, Schwalbach, Germany). After being blocked with TBS containing 5% skimmed milk at room temperature for 1 h, the membrane was incubated with rabbit anti-PDCD4 antibody (1:1000, ab51495, Abcam, Cambridge, UK) and anti-GAPDH antibody (1:2000; ab37168, Abcam, Cambridge, UK), respectively at 4 °C overnight, and then incubated with HRP-conjugated secondary antibodies (1:5000, Beyotime, Shanghai, China) at room temperature for 1 h. GAPDH was used as the internal control. Ultimately, the protein bands were developed by the enhanced chemiluminescence reagent (Beyotime, Shanghai, China).
Cell counting kit-8 (CCK-8) assay
Cells were harvested 24 h after the transfection. A total of 1 × 103 CRC cells was transferred into each well of the 96-well plates and CCK-8 assay was performed every 24 h. Briefly, 10 μL of CCK-8 solution (Sigma, St. Louis, MO, USA) was added into each well at the corresponding time points, and the cells were cultured for another 1 h. The the viability of the cells (indicated by the value of absorbance) was analyzed at a wavelength of 450 nm, using a microplate reader (Potenov, Beijing, China). 4 d later, the proliferation of the cells in each group was plotted, with the absorbance values as the ordinate, and the time as the abscissa.
Transwell assay
Cell migration and invasion assays were performed using Transwell chambers (Corning Corning, NY, USA). In cell migration assay, a total of 5 × 105 cells was suspended in serum-free medium and transferred into the upper chamber of each Transwell insert, while the lower chamber was added with 600 μL of complete medium with 20% FBS. After the culture for 24 h, cells in the upper surface of the filter were removed with cotton swabs and cells remaining on the bottom surface of the filter were fixed with methanol at room temperature for 15 min, followed by being staining with 1% crystal violet at room temperature for 30 min. Finally, stained cells were photographed under a light microscope at × 200 magnification and, and the number of these cells of five randomly selected fields was counted. In cell invasion assay, the filter was pre-coated with diluted Matrigel, and the other procedures were executed as described above.
Flow cytometry
Cell apoptosis was detected by Annexin V-FITC Apoptosis Detection Kit (Sigma, St. Louis, MO, USA). Transfected cells were centrifugated at 5000×g for 5 min at room temperature. Cell pellets were rinsed with PBS and re-suspended in the staining buffer. Then the cells were stained with 5 μL of propodium iodide staning solution in the dark for 30 min at 4 °C and subsequently stained with 5 μL of Annexin V-FITC staining solution for 20 min at room temperature. After that, apoptotic cells were analyzed by a flow cytometer (BD Biosciences, San Jose, CA, USA).
Dual-luciferase reporter assay
The wild type (WT) fragments of circ_0003266 / PDCD4 3′-untranslated region (UTR) containing the predicted binding sites of miR-503-5p, and mutant (MUT) circ_0003266/PDCD4 3’UTR sequences were provided by GenePharma Co., Ltd. (Shanghai, China). The fragments were cloned into the pGL3 Dual-Luciferase miRNA Target Expression vector (Promega, Madison, WI, USA), according to the manufacturer’s protocol. The miR-503-5p mimic or negative control mimic was co-transfected into HEK-293 T cells with the wild-type or mutant reporter vectors. After 48 h, the relative activity of luciferase was determined using the Dual-Luciferase Reporter Assay kit (Promega, Madison, WI, USA) in line with the manufacturer’s instructions.
Statistical analysis
Graphs were generated by GraphPad Prism 8.0 (GraphPad Software, Inc., La Jolla, CA, USA), and statistical analysis was performed with SPSS 22.0 (IBM, Chicago, IL, USA.). Student’s t-test or one-way analysis of variance was adopted for making the comparisons. Pearson correlation analysis was conducted to analyze the correlations between the two indicators. Chi-square test was used to analyze the association between circ_0003266 expression and the clinical characteristics of the patients. P < 0 .05 was considered statistically significant.
Results
Circ_0003266 is lowly expressed in CRC
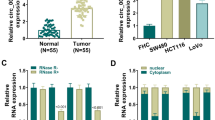
The expression profile of circRNAs in 5 pairs of CRC tissue and normal tissue samples were analyzed by circRNA microarray (GSE142837). Out of 3292 circRNAs, 43 circRNAs (|log2FC| > 1, P < 0.05) were screened out, among which the expression levels of 13 circRNAs were up-regulated and 30 were down-regulated (Fig. 1a-b). In this study, circ_0003266, whose expression was significantly down-regulated, was investigated. CircRNAs have no free ends, showing a longer half-life and resistance to RNase R, compared with liner RNA. To verify the circular structure of circ_0003266, RNase R resistance assay was performed, and it was found that only linear mRNA (GAPDH) expression level was decreased after RNA R treatment, implying that circ_0003266 has a loop structure (Fig. 1c). Next, we found that circ_0003266 expression in CRC tissues was markedly lower than that in adjacent tissues by qRT-PCR (Fig. 1d). Besides, circ_0003266 expression in CRC cell line was significantly down-regulated compared with that in NCM460 cells (Fig. 1e). Furthermore, we analyzed the relationship between the expression of circ_0003266 and the clinicopathological features of CRC patients. The results illustrated that circ_0003266 expression level was negatively correlated with tumor grade and stage of CRC patients (Table 2). These dates suggested that the abnormal down-regulation of circ_0003266 expression might affect CRC progression.
Circ_0003266 was lowly expressed in CRC. a. Volcano plot showed the up-regulated and down-regulated circRNAs in CRC tissues (v.s. non-cancerous tissues) from the analysis of GSE142837. Highly-expressed circRNAs were indicated by “red”, and the lowly-expressed circRNAs were indicated by “green”. b. Heat map showed the down-regulated circRNAs in CRC tissues v.s. non-cancerous tissues from the analysis of GSE142837. c. qRT-PCR was used to detect the expression levels of circ_0003266 and GAPDH after the treatment of RNase R. d. qRT-PCR was used to detect the expression of circ_0003266 in human CRC tissues and adjacent tissues. e. qRT-PCR was used to detect the expression of circ_0003266 in normal colonic epithelial cells (NCM460) and CRC cell lines (HT29, SW480, HCT-116, Lovo, and DLD-1). ***P < 0.001
Circ_0003266 suppresses the malignant phenotypes of CRC cells
Circ_0003266 overexpression plasmid was constructed and transfected into SW480 cells, and si-circ_0003266 was transfected into HCT-116 cells. The transfection efficacy was confirmed by qRT-PCR (Fig. 2a). To further clarify the function of circ_0003266 in CRC, CCK-8, flow cytometry, and Transwell assays were performed, and the results displayed that circ_0003266 overexpression significantly decreased the growth, migration, and invasion of SW480 cells, and expedited the apoptosis, but circ_0003266 silencing functioned oppositely on HCT-116 cells (Fig. 2 b-d). These results showed that circ_0003266 was a tumor suppressor in CRC.
Circ_0003266 inhibited the proliferation and metastasis of CRC cells, and promoted the apoptosis. a. qRT-PCR was employed to detect the expression of circ_0003266 in SW480 and HCT-116 cells transfected with circ_0003266 or two independent siRNAs targeting circ_0003266. b. CCK8 assay was used to detect the effects of circ_0003266 on the proliferation capability of SW480 and HCT-116 cells. c. Transwell assay was used to detect the effects of circ_0003266 on migration and invasion capability of SW480 and HCT-116 cells. d. Flow cytometry was used to detect the effects of circ_0003266 on the apoptosis rate of SW480 and HCT-116 cells. **P < 0.01 and ***P < 0.001
Circ_0003266 negatively regulates miR-503-5p expression
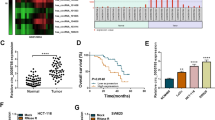
We analyzed the candidate miRNAs with complementary sequences to circ_0003266 using the online CircInteractome database, and found that miR-503-5p was one of the potential targets (Fig. 3a). Dual-luciferase reporter gene assay uncovered that miR-503-5p could decrease the luciferase activity of circ_0003266-WT reporter but had no significant effect on circ_0003266-MUT reporter (Fig. 3b). Besides, qRT-PCR results showed that circ_0003266 overexpression resulted in a significant decrease in miR-503-5p expression in CRC cells, while the depletion of circ_0003266 worked oppositely (Fig. 3c). Besides, miR-503-5p expression in CRC tissues was higher than that in adjacent tissues and negatively correlated with circ_0003266 expression (Fig. 3 d-e). These evidence indicated that miR-503-5p was the target of circ_0003266.
MiR-503-5p was a direct target of circ_0003266. a. Circular RNA Interactome was used to predict the binding site between circ_0003266 and miR-503-5p. b. Dual-luciferase reporter gene assay was used to confirm the relationship between circ_0003266 and miR-503-5p. c. qRT-PCR was used to detect the expression of miR-503-5p in SW480 and HCT-116 cells transfected with NC, circ_0003266 overexpression plasmids, si-NC, or si-circ_0003266. d. qRT-PCR was used to detect the expression of miR-503-5p in human CRC tissues and adjacent tissues. e. Pearson correlation analysis showed the negative relationship between circ_0003266 and miR-503-5p in CRC tissues. ***P < 0.001
MiR-503-5p negatively regulates PDCD4 expression
Through analyzing StarBase databse, TargetScan database, and miRDB database, we found that there was a binding site in PDCD4 3’UTR to miR-503-5p (Fig. 4a), which is consistent with the previous report [14]. Dual-luciferase reporter gene assay indicated that miR-503-5p overexpression markedly repressed the luciferase activity of PDCD4-WT reporter but did not exert an impact on that of PDCD4-MUT reporter (Fig. 4b). MiR-503-5p mimics significantly inhibited PDCD4 mRNA and protein expression levels while miR-503-5p inhibitors worked oppositely (Fig. 4c-d). Meanwhile, there was a negative correlation between the expression levels of miR-503-5p and PDCD4 in CRC tissues (Fig. 4e).
PDCD4 was confirmed as a target gene of miR-503-5p. a. StarBase was used to predict the binding site of miR-503-5p and PDCD4. b. Dual-luciferase reporter gene assay was used to confirm the relationship between miR-503-5p and PDCD4. c-d. qRT-PCR and Western blot were used to detect the expression of PDCD4 in CRC cells after miR-503-5p was modulated. e. Pearson correlation analysis showed the negative relationship between PDCD4 and miR-503-5p in CRC tissues. ***P < 0.001
Circ_0003266 restrains CRC progression via modulating miR-503-5p/PDCD4 axis
To pinpoint whether circ_0003266 exerted its functions via modulating miR-503-5p/PDCD4 pathway, we performed rescue assays. The SW480 cells were divided into four groups: NC group, circ_0003266 overexpression group, circ_0003266 overexpression + miR-503-5p overexpression group, and circ_0003266 overexpression + miR-503-5p overexpression + PDCD4 overexpression group. Western blot assay showed that, circ_0003266 significantly promoted the expression of PDCD4 in SW480 cells, and the co-transfection of miR-503-5p reversed this effects, and the transfection of PDCD4 overexpression plasmids counteracted the effects of miR-503-5p (Fig. 5a). Furthermore, functional assays showed that, circ_0003266 overexpression inhibited the proliferation, migration, and invasion, and promoted the apoptosis of SW480 cells; however, miR-503-5p mimics totally reversed these effects (Fig. 5b-e); additionally, PDCD4 overexpression reversed the effects induced by miR-503-5p overexpression (Fig. 5b-e). These findings demonstrated that circ_0003266 suppressed CRC via modulating miR-503-5p/PDCD4 pathway.
Circ_0003266 inhibited malignant phenotypes of CRC cells by regulating miR-503-5p/PDCD4 axis. a. With transfection, SW480 cells were divided into four groups: NC group, circ_0003266 overexpression group, circ_0003266 overexpression + miR-503-5p overexpression group, and circ_0003266 overexpression + miR-503-5p overexpression + PDCD4 overexpression group, and Western blot assay was used to detect the expression of PDCD4 in SW480 cells. b. CCK8 assay was used to detect the proliferation of SW480 cells after the transfection. c-d. Transwell assay was used to detect the migration and invasion of SW480 cells after the transfection. e. Flow cytometry was used to detect the apoptosis rate of SW480 cells after the transfection. **P < 0.01 and ***P < 0.001
Discussion
CircRNAs are discovered in RNA viruses as early as the in the 1970s, and in recent year, multiple circRNAs are identified in the transcriptome of human cells [15]. Reportedly, circRNAs are more stable and abundant than linear RNA, and circRNAs are mainly located in the cytoplasm and they have miRNA response elements; what’s more, circRNAs have other biological functions such as working as the scaffold in the assembly of protein complexes, regulating alternative splitting, modulating RNA-protein interactions, and so on [15,16,17,18]. CircRNAs are implicated in regulating the pathogenesis of human diseases including diabetes, nervous system diseases, cardiovascular diseases, and cancers, etc. [19]. For example, CirchHipk3 expression is observably raised in CRC tissues and cell lines, and functionally, CirchHipk3 knock-down can markedly impede the growth, migration, and invasion of CRC cells [20]. Circ-ITGA7 inhibits CRC cell proliferation via adsorbing miR-3187-3p and increasing ASXL1 expressions [21]. Here, we found that circ_0003266 expression was significantly down-regulated in CRC. Additionally, circ_0003266 restrained the proliferation and metastatic potential of CRC cells, and expedited the apoptosis. Our results suggested that it could probably be a biomarker and therapy target for CRC.
MiRNAs regulate mRNA expression by inhibiting translation or promoting degradation, and they are important regulators in cancer biology [22]. For example, the decreased expression of miR-4319, as reported, is related to the poor prognosis of CRC patients, and miR-4319 significantly inhibits the proliferation of CRC cells and changes cell cycle distribution by targeting ABTB1 [23]. Reportedly, circRNAs act as miRNAs sponges to regulate tumor progression. For example, circ_0136666 accelerates the multiplication and invasion of CRC cells via miR-136/SH2B1 axis [24]. In this work, we identified miR-503-5p as the target miRNA of circ_003266 by bioinformatics analysis and dual-luciferase reporter gene assay. MiR-503-5p is abnormally expressed in various cancers including hepatocellular carcinoma, ovarian cancer, cervical cancer, and oral squamous cell carcinoma [25,26,27,28]. Besides, miR-503-5p expression in CRC is significantly increased, which expedites the migration and invasion of CRC cells [14]. In this work, we observed that miR-503-5p expression was elevated in CRC tissues, and miR-503-5p promoted proliferation and metastasis of CRC cells, and inhibited the apoptosis, which is consistent with findings of the previous research [14]. Moreover, miR-503-5p could counteract the inhibitory effects of circ_0003266 on CRC procession. These findings suggested that circ_0003266 contributed to the dysregulation of miR-503-5p in CRC, and its function was dependent on miR-503-5p.
PDCD4 is a tumor suppressor, and its expression is frequently down-regulated in various types of cancers [29]. PDCD4 protein is composed of 469 amino acid residues, and PDCD4 binds to eIF4A and restrains its helicase activity [30,31,32]. PDCD4 expression is abnormally down-regulated in CRC, and PDCD4 represses the translation of Sin1 translation via interacting the eIF4A, and inhibits CRC progression [30]. PDCD4 also directly combines with mRNA of c-Myb, Bcl-xL, and XIAP to suppress their translation, thereby inhibiting cell proliferation and promoting apoptosis [32]. Previous studies report that PDCD4 inhibits the progression of several cancer cells, including hepatocellular carcinoma, breast cancer, and melanoma [33,34,35]. Reportedly, miR-503-5p can target PDCD4 [14]. In this study, we further explored the impact of circ_0003266 on PDCD4, the results of which demonstrated that circ_0003266 could positively regulate PDCD4 via adsorbing miR-503-5p.
There are some limitations of the present work. Firstly, our findings are only based on in vitro experiments, and in vivo assays can further confirm the role of circ_0003266 in CRC progression in the future. Secondly, the relationship between circ_0003266 and the prognosis of the CRC patients is still obscure, and survival analysis of more patients with follow-up information should be performed in the future to evaluate the prognostic value of circ_0003266. Lastly, CircInteractome database also predicts other miRNAs, which can probably be regulated by circ_0003266, and whether circ_0003266 could regulate CRC progression via modulating these miRNAs should be explored.
Conclusion
Circ_0003266 is lowly expressed in CRC tissues and cells. Mechanistically, circ_0003266 inhibits CRC progression via modulating PDCD4 expressions by acting as ceRNA of miR-503-5p. The findings of the present study highlight the potential role of circ_0003266 as a tumor suppressor in CRC, which provides a novel therapeutic target for CRC treatment.
Availability of data and materials
The data used to support the findings of this study are available from the corresponding author upon request.
Abbreviations
- CircRNAs:
-
Circular RNAs
- CRC:
-
Colorectal cancer
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
- PDCD4:
-
Programmed cell death 4
- CCK-8:
-
Cell counting kit-8
- ceRNA:
-
Competitive endogenous RNA
- miRNA:
-
MicroRNA
- FBS:
-
Fetal bovine serum
- siRNAs:
-
Small interference RNAs
- PVDF:
-
Polyvinylidene fluoride
- PBS:
-
Phosphate buffered saline
References
Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet (London, England). 2019;394(10207):1467–80.
Geng Y, Zheng X, Hu W, Wang Q, Xu Y, He W, et al. Hsa_circ_0009361 acts as the sponge of miR-582 to suppress colorectal cancer progression by regulating APC2 expression. Clin Sci (London, England : 1979). 2019;133(10):1197–213.
Weinberg BA, Marshall JL, Salem ME. The Growing Challenge of Young Adults With Colorectal Cancer. Oncology (Williston Park, NY). 2017;31(5):381–9.
Yiu AJ, Yiu CY. Biomarkers in colorectal Cancer. Anticancer Res. 2016;36(3):1093–102.
Lech G, Słotwiński R, Słodkowski M, Krasnodębski IW. Colorectal cancer tumour markers and biomarkers: recent therapeutic advances. World J Gastroenterol. 2016;22(5):1745–55.
You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18(4):603–10.
Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell. 2015;58(5):870–85.
Bach DH, Lee SK, Sood AK. Circular RNAs in Cancer. Mol Ther Nucleic Acids. 2019;16:118–29.
Zhu X, Wang X, Wei S, Chen Y, Chen Y, Fan X, et al. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 2017;284(14):2170–82.
Ge J, Jin Y, Lv X, Liao Q, Luo C, Ye G, et al. Expression profiles of circular RNAs in human colorectal cancer based on RNA deep sequencing. J Clin Lab Anal. 2019;33(7):e22952.
Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79.
Wu HB, Huang SS, Lu CG, Tian SD, Chen M. CircAPLP2 regulates the proliferation and metastasis of colorectal cancer by targeting miR-101-3p to activate the notch signalling pathway. Am J Transl Res. 2020;12(6):2554–69.
Zhang L, Dong X, Yan B, Yu W, Shan L. CircAGFG1 drives metastasis and stemness in colorectal cancer by modulating YY1/CTNNB1. Cell Death Dis. 2020;11(7):542.
Li L, Zhang X, Yi Z, Liang X, Yin W, Li S. MiR-503 promotes the migration and invasion of colorectal cancer cells by regulating PDCD4. J BUON. 2018;23(3):579–86.
Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94.
Yin Y, Long J, He Q, Li Y, Liao Y, He P, et al. Emerging roles of circRNA in formation and progression of cancer. J Cancer. 2019;10(21):5015–21.
Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24(2):357–70.
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66.
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–91.
Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9(4):417.
Yang G, Zhang T, Ye J, Yang J, Chen C, Cai S, et al. Circ-ITGA7 sponges miR-3187-3p to upregulate ASXL1, suppressing colorectal cancer proliferation. Cancer Manag Res. 2019;11:6499–509.
Deb B, Uddin A, Chakraborty S. miRNAs and ovarian cancer: an overview. J Cell Physiol. 2018;233(5):3846–54.
Huang L, Zhang Y, Li Z, Zhao X, Xi Z, Chen H, et al. MiR-4319 suppresses colorectal cancer progression by targeting ABTB1. United European Gastroenterol J. 2019;7(4):517–28.
Jin C, Wang A, Liu L, Wang G, Li G. Hsa_circ_0136666 promotes the proliferation and invasion of colorectal cancer through miR-136/SH2B1 axis. J Cell Physiol. 2019;234(5):7247–56.
Jiang SP, Li ZR. MiR-503-5p regulates cell epithelial-to-mesenchymal transition, metastasis and prognosis of hepatocellular carcinoma through inhibiting WEE1. Eur Rev Med Pharmacol Sci. 2019;23(5):2028–37.
Li X, Han X, Yang J, Sun J, Wei P. miR-503-5p inhibits the proliferation of T24 and EJ bladder cancer cells by interfering with the Rb/E2F signaling pathway. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2017;33(10):1360–4.
Ran W, Zeng YH, Ma XJ, Liao P, Liu XL. The Effect of miR-503-5p on the Proliferation, Invasion, Migration and Epithelial Interstitium of Cervical Cancer HeLa Cells via Targeting E2 F3. Sichuan Da Xue Xue Bao Yi Xue Ban. 2020;51(2):178–84.
Han L, Cheng J, Li A. hsa_circ_0072387 suppresses proliferation, metastasis, and glycolysis of Oral squamous cell carcinoma cells by Downregulating miR-503-5p. Cancer Biother Radiopharm. 2020.
Zeng T, Zhang Q, Yu X, Gao X, Qiu Y. Inhibition of cell migration and invasion and promotion of cell apoptosis by overexpression of programmed cell death 4 (PDCD4) in cervical cancer Siha cells. Int J Clin Exp Pathol. 2018;11(9):4676–83.
Wang Q, Zhu J, Wang YW, Dai Y, Wang YL, Wang C, et al. Tumor suppressor Pdcd4 attenuates Sin1 translation to inhibit invasion in colon carcinoma. Oncogene. 2017;36(45):6225–34.
Montero H, Pérez-Gil G, Sampieri CL. Eukaryotic initiation factor 4A (eIF4A) during viral infections. Virus Genes. 2019;55(3):267–73.
Wang Q, Yang HA-O. The role of Pdcd4 in tumour suppression and protein translation. LID. https://doi.org/10.1111/boc.201800014 (1768-322X (Electronic)).
Huang H, Wang X, Wang C, Zhuo L, Luo S, Han S. The miR-93 promotes proliferation by directly targeting PDCD4 in hepatocellular carcinoma. Neoplasma. 2017;64(5):770–7.
Wang Y, Liu Z, Shen J. MicroRNA-421-targeted PDCD4 regulates breast cancer cell proliferation. Int J Mol Med. 2019;43(1):267–75.
Yin Y, Zhao B, Li D, Yin G. Long non-coding RNA CASC15 promotes melanoma progression by epigenetically regulating PDCD4. Cell Biosci. 2018;8:42.
Acknowledgments
We thank Hubei Yican Health Industry Co., Ltd. for its linguistic assistance during the preparation of this manuscript.
Funding
No funding.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: CHW and XQF; Performed the experiments: CHW and XQF; Statistical analysis: HGY, YG, and GSW; Wrote the paper: CHW and XQF. All authors read and approved the final manuscript.
Authors’ information
Not Applicable.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Institutional Ethics Committee of Yichang Central People’s Hospital, and written informed consents were obtained from all patients before the research.
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wen, C., Feng, X., Yuan, H. et al. Circ_0003266 sponges miR-503-5p to suppress colorectal cancer progression via regulating PDCD4 expression. BMC Cancer 21, 284 (2021). https://doi.org/10.1186/s12885-021-07997-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-021-07997-0