Abstract
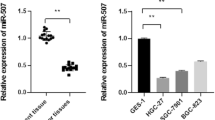
MiR-200a acts as a key role in tumor malignant progression. This work purposed to assess the function of miR-200a in Wilm’s tumor. Based on bioinformatics analysis, the expression, prognostic value and related pathways of miR-200a and CDC7 (a potential downstream molecule of miR-200a) in Wilm’s tumor were analyzed. qRT-PCR was conducted to confirm the miR-200a level in Wilm’s tumor cells. The luciferase reporter assay was carried out to verify the binding of miR-200a to 3′-UTR of CDC7. Then, the impacts of miR-200a and CDC7 on cell viability and apoptosis were measured using CCK-8 and flow cytometry assays. Also, western blot was applied to measure the expression of CDC7 as well as Wnt/β-catenin signaling pathway-related proteins and apoptosis proteins. Herein, we revealed that miR-200a was lowly expressed in Wilm’s tumor tissues and cells and the low miR-200a expression is closely bound up with death and poor outcomes. Moreover, miR-200a directly targeted and inhibited CDC7 in Wilm’s tumor cells. Biological function experiments illustrated that overexpression of miR-200a reduced the viability and elevated the apoptosis of Wilm’s tumor cells, while overexpression of CDC7 reversed the inhibitory impact of miR-200a on cell viability and the promoting impact of miR-200a on cell apoptosis. Besides, we revealed that miR-200a/CDC7 axis can decrease the expression of β-Catenin, Cyclin D1 and C-Myc as well as the phosphorylation of GSK-3β, thus inhibiting the Wnt/β-catenin signaling pathway. Furthermore, blocking the Wnt/β-catenin signaling pathway caused an increase on cell apoptosis, while overexpression of CDC7 can reverse these impacts. Collectively, miR-200a/CDC7 axis involved in regulating the malignant phenotype of Wilm’s tumor through Wnt/β-catenin signaling pathway, which provides a theoretical basis for targeted molecular therapy of Wilm’s tumor.






Similar content being viewed by others
References
Treger TD, Chowdhury T, Pritchard-Jones K, Behjati S (2019) The genetic changes of Wilms tumour. Nat Rev Nephrol 15:240–251
Varan A (2008) Wilms’ tumor in children: an overview. Nephron Clin Pract 108:c83-90
Davidoff AM (2012) Wilms tumor. Adv Pediatr 59:247–267
Dome JS, Graf N, Geller JI, Fernandez CV, Mullen EA, Spreafico F et al (2015) Advances in Wilms tumor treatment and biology: progress through international collaboration. J Clin Oncol 33:2999–3007
Sonn G, Shortliffe LM (2008) Management of Wilms tumor: current standard of care. Nat Clin Pract Urol 5:551–560
Dix DB, Seibel NL, Chi YY, Khanna G, Gratias E, Anderson JR et al (2018) Treatment of stage IV favorable histology Wilms tumor with lung metastases: a report from the children’s oncology group AREN0533 study. J Clin Oncol 36:1564–1570
Warmann SW, Nourkami N, Frühwald M, Leuschner I, Schenk JP, Fuchs J et al (2012) Primary lung metastases in pediatric malignant non-Wilms renal tumors: data from SIOP 93–01/GPOH and SIOP 2001/GPOH. Klin Padiatr 224:148–152
Nishinakamura R, Uchiyama Y, Sakaguchi M, Fujimura S (2011) Nephron progenitors in the metanephric mesenchyme. Pediatr Nephrol 26:1463–1467
Cone EB, Dalton SS, Van Noord M, Tracy ET, Rice HE, Routh JC (2016) Biomarkers for Wilms tumor: a systematic review. J Urol 196:1530–1535
Balzano F, Cruciani S, Basoli V, Santaniello S, Facchin F, Ventura C et al (2018) MiR200 and miR302: two big families influencing stem cell behavior. Molecules 23(2):282
Rupaimoole R, Slack FJ (2017) MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 16:203–222
Hosseinahli N, Aghapour M, Duijf PHG, Baradaran B (2018) Treating cancer with microRNA replacement therapy: a literature review. J Cell Physiol 233:5574–5588
Tiwari A, Mukherjee B, Dixit M (2018) MicroRNA key to angiogenesis regulation: miRNA biology and therapy. Curr Cancer Drug Targets 18:266–277
Liu H, Lei C, He Q, Pan Z, Xiao D, Tao Y (2018) Nuclear functions of mammalian microRNAs in gene regulation, immunity and cancer. Mol Cancer 17:64
Pasquinelli AE, Hunter S, Bracht J (2005) MicroRNAs: a developing story. Curr Opin Genet Dev 15:200–205
Sellitti DF, Doi SQ (2015) MicroRNAs in renal cell carcinoma. MicroRNA 4:26–35
Huang GL, Sun J, Lu Y, Liu Y, Cao H, Zhang H et al (2019) MiR-200 family and cancer: from a meta-analysis view. Mol Aspects Med 70:57–71
O’brien SJ, Carter JV, Burton JF, Oxford BG, Schmidt MN, Hallion JC et al (2018) The role of the miR-200 family in epithelial-mesenchymal transition in colorectal cancer: a systematic review. Int J Cancer 142:2501–2511
Koutsaki M, Libra M, Spandidos DA, Zaravinos A (2017) The miR-200 family in ovarian cancer. Oncotarget 8:66629–66640
Liu C, Hu W, Li LL, Wang YX, Zhou Q, Zhang F et al (2018) Roles of miR-200 family members in lung cancer: more than tumor suppressors. Future Oncol 14:2875–2886
Liu P, Chen S, Huang Y, Xu S, Song H, Zhang W et al (2020) LINC00667 promotes Wilms’ tumor metastasis and stemness by sponging miR-200b/c/429 family to regulate IKK-β. Cell Biol Int 44:1382–1393
Zhu H, Zheng T, Yu J, Zhou L, Wang L (2018) LncRNA XIST accelerates cervical cancer progression via upregulating Fus through competitively binding with miR-200a. Biomed Pharmacother 105:789–797
Zou Q, Zhou E, Xu F, Zhang D, Yi W, Yao J (2018) A TP73-AS1/miR-200a/ZEB1 regulating loop promotes breast cancer cell invasion and migration. J Cell Biochem 119:2189–2199
Feng C, Zhao Y, Li Y, Zhang T, Ma Y, Liu Y (2019) LncRNA MALAT1 promotes lung cancer proliferation and gefitinib resistance by acting as a miR-200a sponge. Arch Bronconeumol 55:627–633
Guo H, Ingolia NT, Weissman JS, Bartel DP (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466:835–840
Moretti F, D’antona P, Finardi E, Barbetta M, Dominioni L, Poli A et al (2017) Systematic review and critique of circulating miRNAs as biomarkers of stage I-II non-small cell lung cancer. Oncotarget 8:94980–94996
Ito S, Taniyami C, Arai N, Masai H (2008) Cdc7 as a potential new target for cancer therapy. Drug News Perspect 21:481–488
Kulkarni AA, Kingsbury SR, Tudzarova S, Hong HK, Loddo M, Rashid M et al (2009) Cdc7 kinase is a predictor of survival and a novel therapeutic target in epithelial ovarian carcinoma. Clin Cancer Res 15:2417–2425
Clarke LE, Fountaine TJ, Hennessy J, Bruggeman RD, Clarke JT, Mauger DT et al (2009) Cdc7 expression in melanomas, Spitz tumors and melanocytic nevi. J Cutan Pathol 36:433–438
Rodriguez-Acebes S, Proctor I, Loddo M, Wollenschlaeger A, Rashid M, Falzon M et al (2010) Targeting DNA replication before it starts: Cdc7 as a therapeutic target in p53-mutant breast cancers. Am J Pathol 177:2034–2045
Bonte D, Lindvall C, Liu H, Dykema K, Furge K, Weinreich M (2008) Cdc7-Dbf4 kinase overexpression in multiple cancers and tumor cell lines is correlated with p53 inactivation. Neoplasia 10:920–931
Liu YP, Chen WD, Li WN, Zhang M (2019) Overexpression of FNDC1 relates to poor prognosis and its knockdown impairs cell invasion and migration in gastric cancer. Technol Cancer Res Treat 18:1533033819869928
Vijayakumar S, Liu G, Rus IA, Yao S, Chen Y, Akiri G et al (2011) High-frequency canonical Wnt activation in multiple sarcoma subtypes drives proliferation through a TCF/β-catenin target gene, CDC25A. Cancer Cell 19:601–612
Li A, Omura N, Hong SM, Vincent A, Walter K, Griffith M et al (2010) Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer Res 70:5226–5237
Elson-Schwab I, Lorentzen A, Marshall CJ (2010) MicroRNA-200 family members differentially regulate morphological plasticity and mode of melanoma cell invasion. PLoS ONE 5(10):e13176
Wu Q, Lu RL, Li JX, Rong LJ (2017) MiR-200a and miR-200b target PTEN to regulate the endometrial cancer cell growth in vitro. Asian Pac J Trop Med 10:498–502
Jia C, Zhang Y, Xie Y, Ren Y, Zhang H, Zhou Y et al (2019) miR-200a-3p plays tumor suppressor roles in gastric cancer cells by targeting KLF12. Artif Cells Nanomed Biotechnol 47:3697–3703
Xia H, Ng SS, Jiang S, Cheung WK, Sze J, Bian XW et al (2010) miR-200a-mediated downregulation of ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma cell growth, migration and invasion. Biochem Biophys Res Commun 391:535–541
Braun J, Hoang-Vu C, Dralle H, Hüttelmaier S (2010) Downregulation of microRNAs directs the EMT and invasive potential of anaplastic thyroid carcinomas. Oncogene 29:4237–4244
Saydam O, Shen Y, Würdinger T, Senol O, Boke E, James MF et al (2009) Downregulated microRNA-200a in meningiomas promotes tumor growth by reducing E-cadherin and activating the Wnt/beta-catenin signaling pathway. Mol Cell Biol 29:5923–5940
Guo T, Zhang Y, Qu X, Che X, Li C, Fan Y et al (2018) miR-200a enhances TRAIL-induced apoptosis in gastric cancer cells by targeting A20. Cell Biol Int 42:506–514
Guo B, Romero J, Kim BJ, Lee H (2005) High levels of Cdc7 and Dbf4 proteins can arrest cell-cycle progression. Eur J Cell Biol 84:927–938
Kitamura R, Fukatsu R, Kakusho N, Cho YS, Taniyama C, Yamazaki S et al (2011) Molecular mechanism of activation of human Cdc7 kinase: bipartite interaction with Dbf4/activator of S phase kinase (ASK) activation subunit stimulates ATP binding and substrate recognition. J Biol Chem 286:23031–23043
Suzuki T, Tsuzuku J, Hayashi A, Shiomi Y, Iwanari H, Mochizuki Y et al (2012) Inhibition of DNA damage-induced apoptosis through Cdc7-mediated stabilization of Tob. J Biol Chem 287:40256–40265
Zhan T, Rindtorff N, Boutros M (2017) Wnt signaling in cancer. Oncogene 36:1461–1473
Nusse R, Clevers H (2017) Wnt/β-Catenin signaling, disease, and emerging therapeutic modalities. Cell 169:985–999
Ni D, Liu J, Hu Y, Liu Y, Gu Y, Zhou Q et al (2019) A1CF-Axin2 signal axis regulates apoptosis and migration in Wilms tumor-derived cells through Wnt/β-catenin pathway. In Vitro Cell Dev Biol Anim 55:252–259
Zhu KR, Sun QF, Zhang YQ (2019) Long non-coding RNA LINP1 induces tumorigenesis of Wilms’ tumor by affecting Wnt/β-catenin signaling pathway. Eur Rev Med Pharmacol Sci 23:5691–5698
Tian J, He H, Lei G (2014) Wnt/β-catenin pathway in bone cancers. Tumour Biol 35:9439–9445
Behrens J, Von Kries JP, Kühl M, Bruhn L, Wedlich D, Grosschedl R et al (1996) Functional interaction of beta-catenin with the transcription factor LEF-1. Nature 382:638–642
He TC, Sparks AB, Rago C, Hermeking H, Zawel L, Da Costa LT et al (1998) Identification of c-MYC as a target of the APC pathway. Science 281:1509–1512
Schuhmacher M, Eick D (2013) Dose-dependent regulation of target gene expression and cell proliferation by c-Myc levels. Transcription 4:192–197
Pelengaris S, Khan M, Evan G (2002) c-MYC: more than just a matter of life and death. Nat Rev Cancer 2:764–776
Yu J, Wang X, Lu Q, Wang J, Li L, Liao X et al (2018) Extracellular 5’-nucleotidase (CD73) promotes human breast cancer cells growth through AKT/GSK-3β/β-catenin/cyclinD1 signaling pathway. Int J Cancer 142:959–967
Gopalakrishnan N, Saravanakumar M, Madankumar P, Thiyagu M, Devaraj H (2014) Colocalization of β-catenin with Notch intracellular domain in colon cancer: a possible role of Notch1 signaling in activation of CyclinD1-mediated cell proliferation. Mol Cell Biochem 396:281–293
Wu J, Lv S, An J, Lu C (2015) Pre-miR-149 rs71428439 polymorphism is associated with increased cancer risk and AKT1/cyclinD1 signaling in hepatocellular carcinoma. Int J Clin Exp Med 8:13628–13633
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liang, XL., Wang, YL. & Wang, PR. MiR-200a with CDC7 as a direct target declines cell viability and promotes cell apoptosis in Wilm’s tumor via Wnt/β-catenin signaling pathway. Mol Cell Biochem 476, 2409–2420 (2021). https://doi.org/10.1007/s11010-021-04090-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-021-04090-9