Abstract
MicroRNAs (miRNAs) are endogenous non-coding RNAs that contain approximately 22 nucleotides. They serve as key regulators in various biological processes and their dysregulation is implicated in many diseases including cancer and autoimmune disorders. It has been well established that the maturation of miRNAs occurs in the cytoplasm and miRNAs exert post-transcriptional gene silencing (PTGS) via RNA-induced silencing complex (RISC) pathway in the cytoplasm. However, numerous studies reaffirm the existence of mature miRNA in the nucleus, and nucleus-cytoplasm transport mechanism has also been illustrated. Moreover, active regulatory functions of nuclear miRNAs were found including PTGS, transcriptional gene silencing (TGS), and transcriptional gene activation (TGA), in which miRNAs bind nascent RNA transcripts, gene promoter regions or enhancer regions and exert further effects via epigenetic pathways. Based on existing interaction rules, some miRNA binding sites prediction software tools are developed, which are evaluated in this article. In addition, we attempt to explore and review the nuclear functions of miRNA in immunity, tumorigenesis and invasiveness of tumor. As a non-canonical aspect of miRNA action, nuclear miRNAs supplement miRNA regulatory networks and could be applied in miRNA based therapies.
Similar content being viewed by others
MicroRNA (miRNA) is a group of small non-coding RNA that plays significant roles in multiple metabolic processes. Since its discovery in 1993 [1], numerous studies have postulated and established a set of theories concerning miRNA biogenesis and functions, with cross-species researches initially focusing on translational repression in cytoplasm. After transcription, cleavage, and processing, mature miRNA is transported from the nucleus to cytoplasm to be loaded into RNA induced silencing complex (RISC). MiRNA base-pairs with the mRNA, mediating mRNA decay or detachment of ribosomes. In addition to potent inhibitory functions in the cytoplasm, in 2008, research demonstrated that the introduction of miR-373 and its precursor (pre-miR-373) induced the expression of cold-shock domain-containing protein C2 (CSDC2) and E-cadherin [2]. This is attributed to the sequence complementarity of miR-373 and promoters of those genes. In 2004, another member of small non-coding RNA, small interfering RNA (siRNA) was observed to inhibit the transcription of elongation factor 1alpha (EF1A) though promoter interaction [3]. Following these, research interest in promoter-targeting siRNA has increased substantially [4]. With the introduction of newly developed techniques like microarray and RNA-seq, numerous mature miRNAs were found enriched in nucleus, which demonstrates that nuclear miRNAs are more prevalent than what we had thought. Meanwhile, several models elaborating transcriptional gene silencing (TGS) and transcriptional gene activation (TGA) were established involving promoter interaction, non-coding RNA binding, RNA-DNA triplex and enhancer interaction. In addition, recently, several proteins that mediate nucleus-cytoplasm shuttling of key participants in RNA interference (RNAi) were confirmed [5].
This review summarizes nuclear miRNA’s evidence of existence, prevailing models of nuclear miRNA functional mechanisms, practical prediction software tools and associated applications in immunity and cancer. Generally, although different in origin and maturation processes, mature miRNA and siRNA are chemically the same and function similarly when loaded into RISC [6]. Hereby, several researches utilizing siRNA are quoted as analogous examples.
Canonical biogenesis and functions of MicroRNAs
Canonically, the biogenesis of miRNA is a multi-step process involving both nuclear and cytoplasmic machineries. First, the miRNA gene is transcribed by RNA polymerase II to produce a long pri-miRNA [7], which is then bound to the micro-processing complex consisting of RNA binding protein DGCR8 and the RNase III Drosha [8]. Drosha initiates miRNA maturation by cleaving the pri-miRNA to form the hairpin-structured pre-miRNA [9]. Then the pre-miRNA produced is exported to the cytoplasm via Exportin 5 where maturation is completed [10]. In the cytoplasm, pre-miRNA is recognized with its characteristic 2-nt 3′ overhang by Dicer [11], which cleaves off the terminal loop of the hairpin of the pre-miRNA and generates a miRNA duplex. The protein TRBP stabilizes Dicer and chaperones it with dicing functions [12]. Finally, the miRNA duplex is accommodated and unwound by cytoplasmic Argonaute protein (Ago). One strand is retained to form the functional miRNA-induced silencing complex (miRISC), while the other strand is degraded [13].
Post-transcriptional gene silencing (PTGS) in cytoplasm is the classic function mediated by miRNA via miRISC. For example, miR-139-5p and miR-144 are able to reduce the expression of TET2 and TET3 on both mRNA and protein level [14]. The first step of PTGS is recognition. Some basic principles of base-paring include canonical Watson-Crick A-U, G-C pairing and non-canonical G-U pairing. There is a special sequence on the target mRNA for miRNA recognition and binding called miRNA response element (MRE). The MRE is mostly located at the 3’-UTR of the mRNA, just like TET2 and TET3 mRNA [14]. But some studies suggest that it also occurs in 5’-UTR and even in the protein-coding sequences [15,16,17,18]. For mammals, the base-pairing is always imperfect. One example is the base-pairing between let-7 and lin-41. There are two MREs in lin-41, nevertheless, neither of which has perfect complementarity with the 5′-end of let-70 [15]. In addition, recent studies pointed out that translation will be blocked via three pathways: (i) Deadenylation and degradation mediated by CAF1/CCR4/NOT1 complex; (ii) 5′-decapping facilitated by Dcp1/2 decapping complex; (iii) Ribosome detachment from target mRNA (Fig. 1).
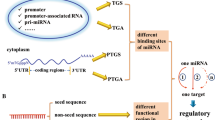
MiRNA biogenesis, function in cytoplasm and nucleus-cytoplasm transport. The biogenesis of miRNA takes several steps. First, pri-miRNA is transcribed via RNA pol II and then cleaved into pre-miRNA by Drosha with DGCR8. Guided by Exportin 5, pre-miRNA is exported to the cytoplasm, where Dicer cuts the stem-loop structure into a double-stranded RNA. The double-stranded RNA formed is then loaded onto Ago 2. One is the mature miRNA, also known as the guide strand. The other is the passenger RNA, which is quickly degraded. Mature miRNA functions both in the cytoplasm and in the nucleus. Core RISC, the major cellular post-transcriptional silencing machinery, is composed of miRNA, Ago 2 and trinucleotide repeat containing 6 (TNRC6). MiRNA recognizes and binds the miRNA response element (MRE) on the mRNA. In the cytoplasm, there are three pathways for miRNAs to exert post-transcriptional silencing: 1) Ago2 recruits TNRC6, which then recruits CCR4-NOT deadenylase complex. This complex leads to deadenylation and degradation of mRNA. 2) TNRC6 recruits Dcp 1/2 decapping complex which cleaves 5′ cap of mRNA and reduces mRNA stability. 3) With binding of Ago 2, mRNA is rendered inaccessible for ribosome attachment and function, which inhibits the translation process. Meanwhile, Ago 2 with mature miRNA is imported into the nucleus via Importin 8 and TNRC6 via Importin β. In the nucleus, RISC is assembled and functions in the transcriptional or post-transcriptional pathway. The nuclear RISC can be transported into cytoplasm via Exportin1 (also referred to as XPO1 or chromosomal region maintenance 1 (CRM1)), which makes transport between the cytoplasm and the nucleus bidirectional. RNAP II, RNA polymerase II; TGS, transcriptional gene silencing; TGA, transcriptional gene activation; PTGS, post-transcriptional gene silencing; Ago2, Argonaute 2; TNRC6, trinucleotide repeat containing 6; TRBP, TAR RNA binding protein; MRE, microRNA response element
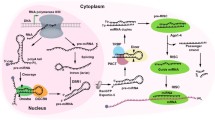
MicroRNA-promoter interaction mechanisms. There are three models for microRNA-promoter interaction, all of which underscore the importance of Ago protein, though there are some conflicting views on whether Ago1 or Ago2 is involved. The resultant interaction effect can be either transcriptional activation or transcriptional suppression. In most cases, the RNA-DNA and the RNA-RNA hybrid model rely on histone modifiers and the recruitment of RNA polymerase II, while whether microRNA can mediate DNA methylation remains dubious. Moreover, in the RNA-DNA-DNA triplex model, the interaction is not related to epigenetic modification. a In the RNA-DNA model, microRNA-Ago complex directly targets to one of the DNA strands which always contains the TATA box motif or transcription factor binding sites when this region is in open configuration for transcription. Then some transcription factors and/or epigenetic modifiers are recruited to the promoter region leading to RNA polymerase II recruitment and/or epigenetic modification. b The RNA-RNA model is related to the non-coding transcripts derived from the promoter region. Either sense or antisense transcript can be a target for microRNA-Ago complex, serving as a scaffold for microRNA-Ago complex and recruiting the histone modifiers and/or transcription factors. c In the RNA-DNA triplex model, microRNAs form triple-helical structures with DNA, altering the topography of the chromatin, which is rendered either more accessible or inaccessible to distinct transcription factors. TF, transcription factor; ncRNA, non-coding RNA; RNA pol II, RNA polymerase II; TSS, transcription start site; Ago, Argonaute 1 or Argonaute 2; Histone modifiers, such as histone methyltransferase enhancer of zeste homolog 2 (EZH2) and euchromatic histone lysine methyltransferase 2 (EHMT2)
Evidence of MicroRNA in the nucleus
Accumulating evidence suggests that there are miRNAs in the nucleus, though it is putative that miRNA remains in the cytoplasm after its biogenesis [19,20,21,22,23,24]. Since microRNA was first identified in Hela cell nucleus [25], the existence of nuclear mature miRNAs has been further supported by several studies differentiating microRNA profiles into cytoplasmic and nuclear fractions. With high-throughput profiling technologies such as microarray and deep sequencing, hundreds of nuclei-enriched microRNAs have been identified in a variety of cell lines (Table 1) [26,27,28,29,30,31,32]. Some of these results are reaffirmed with Northern Blot, RT-qPCR, RT-PCR and in situ hybridization (ISH) to eliminate precursor signals. It is interesting to note that while nuclear localization of microRNA is a relatively general phenomenon, the microRNA profile in the nucleus varies across tissue types. For example, the nuclear enrichment of human miR-29b has been reported in HeLa cells [33] but not in other cell lines [28]. Notably, the miRNA nuclear-cytoplasmic ratio is identified as a significant feature to distinguish three different breast cancer cell lines [31]. In addition, a web-accessible database RNALocate (http://www.rna-society.org/rnalocate/) provides RNA subcellular localization information which was manually obtained from articles published in the PubMed database before May 2016. Some miRNAs with a nuclear localization can be found in RNALocate [34].
Shuttle pathways of RISC components
As with nuclear RNA interference [35], many RISC components have been identified that are functional in the nucleus, including Argonaute 2 (Ago2) and trinucleotide repeat containing 6 (TNRC6, also known as GW182) [36,37,38]. Importin 8, a member of karyopherin β family, has been proven to be the mediator in the nuclear import process of Ago2. Specifically, Ago2 can only be transported when loaded with miRNA [39, 40]. The subcellular distribution of Ago2 varies across cell and tissue. For example, the nuclei of Hela and HaCaT cells present with a minimal level of Ago2 [41]. With both nuclear localization signal (NLS) and nuclear export signal (NES), TNRC6A can be transported from nucleus to cytoplasm with the assistance of Exportin 1 (XPO1, also referred to as CRM1) [42] and inversely with Importin α/β [43, 44].
The nuclear RISC is quite different from its cytoplasmic counterpart: under fluorescence correlation and cross-correlation spectroscopy, RISC presents as an approximately 158 kDa complex in the nucleus whereas 20-fold larger complex of nearly 3 MDa in the cytoplasm [45].When imported into the nucleus, cytoplasmic cofactors such as Dicer and TRBP are not attached to the RISC [45]. Consistently, recent research demonstrated that Ago2 and TNRC6, the core components of RNAi, are conserved in the nucleus and cytoplasm [46].
Given that miRNA is colocalized with Argonaute 2 (Ago2) and TNRC6 in the nucleus and cytoplasm, it is hypothetical that RISC associated proteins serve as not only the executors of RNAi but also agents in the miRNA nuclear import [42] (Fig. 1). Ago2 loaded with miRNA can be transported into nucleus with the mediation of Importin 8, while TNRC6 can be independently transported via Importin α/β. Ago2 and TNRC bind together in the nucleus carrying miRNA, forming a complex, which can be exported via XPO1. This explains why miRNA accumulates in the nucleus with reduction of XPO1 in the previous study [47].
Functions of nuclear MicroRNAs
Regulation of RNAs
It has long been observed that nuclear RISC could mediate post-transcriptional gene silencing (PTGS) with miRNA in the nucleus, as well as in cytoplasm. In such a pathway, other endonuclear non-coding RNAs or miRNA precursors could be degraded. In 2005, researchers efficiently degraded nuclear-localized 7SK snRNA with transfection of perfectly complementary siRNA [35]. Long non-coding RNA (lncRNA), a group of non-coding RNAs over 200 nucleotide in length, is also demonstrated to be subject to endonuclear PTGS control. For example, metastasis associated lung adenocarcinoma transcript 1 (MALAT1) is a target of miR-9 in an Ago2-dependent manner in the nucleus [48]. In the nucleus, the maturation of miR-15a/16–1 is inhibited by miR-709 through binding between pri-miR-15a/16–1 and miR-709 [49], which suggests that expression and maturation of one miRNA could be subject to another miRNA.
Transcriptional regulation by interaction with promoters
There are three models for MicroRNA-promoter interaction based on observation of siRNA directed transcriptional regulation [17, 50] (Fig. 2). All of these three models are associated with the Ago protein, although there are some contradictory results as to whether Ago1 or Ago2 is involved [51]. The regulation can be either activation or suppression, and which mode of action occurs is sensitive to the location of target region (such as TATA box motif and CpG island region) and epigenetic status (such as DNA methylation) of the promoter [52]. Moreover, the target region can be far away from the transcription initiation site. For example, the most effective small activating RNA targeting site is located to 1611-bp upstream of the transcription start site (TSS) [53]. Some histone modifiers have been proven to be recruited to the promoter region. Histone methyltransferase euchromatic histone lysine methyltransferase 2 (EHMT2), which suppresses the expression of fumarate hydratase in nasopharyngeal carcinoma [54], is recruited with enhancer of zeste homolog 2 (EZH2) by miR-584-3p in an Ago2-dependent manner to reduce matrix metalloproteinase 14 (MMP-14) expression in gastric cancer [55]. A recently published study indicated that the genome targeting RNA-induced transcriptional activation complex comprises at least three elements: small RNA-loaded Ago2, RHA (a nuclear DNA helicase II), and CTR9 (a component of PAF1 complex that is involved in transcription initiation and elongation). The complex which targets to the p21 promoter interacts with RNA polymerase II to stimulate transcription initiation and induce mRNA elongation, accompanied by ubiquitination of histone H2B in the p21 gene. This ubiquitination is regarded as a prerequisite for H3K4 methylation and acetylation and followed by histone modification [56]. However, along with the complexity of the epigenetic modifications involved in the regulation process [35], whether miRNA can mediate DNA methylation remains dubious [3, 51, 57].
RNA-RNA model
In this model, the microRNA-Ago complex directly targets to noncoding transcripts (either sense or antisense) and serves as a molecular scaffold to recruit additional epigenetic factors, altering epigenetic modifications (such as H3K4me3 and H3K27me3) [58,59,60,61]. This complex can also induce enrichment of RNA polymerase II at the promoter region [58, 59, 62].
The transcriptional regulation of siRNA can be different in different cellular contexts. For example, the expression of progesterone receptor (PR) is low in MCF7 breast cancer cells, but higher in T47D cells. Promoter-directed siRNA which activates PR expression in MCF7 cells has no effect or even inhibits PR expression in T47D cells [59, 63]. Some antisense transcripts exist within the PR promoter. Solely reduction of the antisense transcript disrupts the effect of siRNA, but basal expression of PR is not changed in MCF7 cells. They also observed the direct interaction between siRNAs and the antisense transcript by biotin labeling. However, the mechanism through which siRNAs are able to activate or inhibit gene expression is poorly understood [59].
However, another model is suggested for miRNA-promoter interaction based on the data of bidirectional gene regulation [62]. At steady state, endogenous p21 contains comparable levels of both sense and antisense transcripts. Decreased antisense RNA induces loss of H3K27me3 and Ago1 at the p21 promoter, thus downregulating the promoter-associated RNA and upregulating the sense RNA. Decreased sense RNA can also alter the epigenetic modifications.
By targeting the promoter, miRNA can also activate another gene that is located at the upstream of the promoter. The underlying mechanism is related to the noncoding RNA overlapping the promoter and gene looping rendering two linearly distant promoter region spatially proximal [60, 64].
RNA-DNA hybrid model
MiRNAs guide Ago protein to promoter targets such as TATA-box motif or the regions associated with transcript factors. Then, transcription preinitiation complex (PIC) is formed, altering histone epigenetic modification as well as binding of transcript factors. Small RNAs might interact with single-stranded DNA while the DNA double helix unwinds during transcription initiation [53, 55, 65,66,67].
RNA-DNA triplex model
In this model, purine or pyrimidine rich (> 75%) miRNAs form triple-helical structures with purine-rich duplex DNA via Hoogsteen or reverse Hoogsteen interaction in the major groove of the duplex DNA. This interaction may alter the DNA topography and allow binding of transcription factors, resulting in transcriptional activation or suppression [68, 69]. Paugh et al. developed an algorithm (Trident) to search for potential triplex-formation sites. Meanwhile, they detected a transient interaction between hsa-miR-5p (a miRNA rich in purine content) with a double-stranded DNA (an identified Hoogsteen binding site screened in genome) in vitro via some methods that are more sensitive and effective in detection of stable triplexes than electrophoretic mobility shift assay (EMSA). Moreover, in primary leukemia cells, largely, the expression of miRNAs that are predicted by Trident are positively correlated with the expression of their target [68]. However, as few evidence exists for triplex formation of miRNAs in vivo [69, 70], this model still needs to be validated upon further study.
MicroRNAs serve as enhancer trigger in the nucleus
Enhancers are genomic cis-regulatory elements able to upregulate gene transcription. Some markers of enhancer including H3K27ac are found in the microRNA genes (MIRs), which means some MIRs and enhancers overlap. Some miRNAs, including miR-26a-1, miR-339, miR-3179, miR-24-1, miR-24-2, had been proven to be able to induce expression of neighboring genes [71]. For example, miR-26a-1 gene is surrounded by protein-coding genes ITGA9, CTDSPL, VILL and PLCD1. The overexpression of miR-26a-1 will lead to transcriptional activation of ITGA9 and VILL in HEK293T. The activation is disrupted when the seed region of miRNA is deleted or mutated or when the enhancer locus is deleted, which suggests that this function relies on miRNA-enhancer base-pairing. Another miRNA, miR-24-1, also increases the expression of its neighboring genes, FBP1 and FANC. Increased miR-24-1 will lead to enrichment of RNA polymerase II, p300/CBP, enhancer RNAs, all of which indicate active regulatory functions. Interestingly, ChIP-qPCR demonstrates that there is Ago2 at the enhancer locus. Furthermore, no transcriptional activation of neighboring genes of miR-24-1 will occur if Ago2 is knocked down [71]. So, it could be argued that enhancer activation induced by miRNAs requires Ago2 to function directly at the locus or carry mature miRNA from cytoplasm to the nucleus.
Similarities and differences between the nuclear and Cytoplasmic functions of MicroRNA
In accordance to those high-throughput profiling results, a large part of miRNAs shuttle between nucleus and cytoplasm, which indicates that they may have both nuclear and cytoplasmic functions. Current knowledge of similarities and differences between miRNA nuclear and cytoplasmic functions are presented as follows (Table 2).
Both functions are potent in regulation of multiple processes and mediators of miRNA nuclear and cytoplasmic functions partially overlap. Though there is evidence indicating that nuclear RISC is smaller than cytoplasmic RISC [45]. The nuclear localization of Ago2 and TNRC6, as the core RISC, have been proven. Therefore, PTGS can be induced both in nucleus and cytoplasm.
Other than similarities, however, differences are obvious between cytoplasmic and nuclear miRNAs due to their different localization, targets and mechanisms. MiRNAs in cytoplasm only have the access to mature mRNA but not DNA. Hereby, cytoplasmic functions involve only post-transcriptional modification via miRNA-mRNA hybrid interaction. Translation repression is mediated though cleavage, de-adenylation, 5′-decapping or ribosome detachment. Whereas, miRNA in nucleus have the chance to interact with DNA and RNA in nucleoplasm. Therefore, both transcriptional and post-transcriptional regulation exist in nucleus. However, nuclear miRNA shares the same post-transcriptional gene silencing (PTGS) mechanism with cytoplasmic miRNA. Targets in nucleus are endonuclear RNAs including non-coding RNAs and precursors of miRNAs. Transcription regulation is mainly mediated via miRNA-promoter interaction. Three miRNA-promoter interaction models are, as reviewed above, RNA-RNA hybrid, RNA-DNA hybrid and RNA-DNA triplex. For the RNA-RNA hybrid model, miRNA-Ago complex targets non-coding transcripts and recruits epigenetic factors to alter epigenetic modification at the promoter region [59]. MiRNA can also directly target promoter facilitated by transcription preinitiation complex to alter histone modification [56]. In the RNA-DNA triplex model, miRNA forms triple-helical structure with DNA to activate or suppress transcription via altering DNA topography [68, 69]. Recently, enhancer associated mechanism has also been discovered [71]. In conclusion, there are differences of target and mechanisms between nuclear and cytoplasmic functions.
Nuclear MicroRNA target prediction software tools
Computational prediction of nuclear miRNA target is a critical initial step. Some traditional tools were exploited, while some new ones were developed based on new mechanisms. Target prediction is generally based on sequential characteristics. There are some differences between nuclear and cytoplasmic miRNA target prediction. Here, we summarize some principles of nuclear target recognition and some public software tools.
Principles of target recognition
The basic principle of microRNA target prediction algorithms is the complement of 5′ end of miRNA and target sequence. As the validation of experimental miRNA-mRNA interaction, several empirical miRNA seed sequence models have been proposed, such as nucleotides from position 2 to 8 in the 5′ end of the miRNA [72]. There is possibly an analogous seed model to be exploited for nuclear miRNA target prediction, as some study shows that the mutation or deletion of seed sequence disrupt the activity of nuclear miRNAs [71, 73].
Additionally, the thermodynamic stability will be measured to evaluate the binding between miRNA and target. A lower free energy indicate a more stable interaction. A widely used software tool to calculate the binding energy is RNAhybrid [74]. There are diverse calculation formula for thermodynamic stability of miRNA-mRNA, miRNA-ssDNA and miRNA-dsDNA interaction, so different algorithms will be used for cytoplasmic miRNA and nuclear miRNA target predicting [68].
Another valuable trait of sequences is their evolutionary conservation. There is more possibility a sequence is vital if it is more conserved. Conserved regions in promoters largely overlap open chromatin regions and TF binding sites [75]. In some prediction software tools, conservation of seed sequences will be measured to prevent some false positive results. However, sequence conservation of the promoter is generally lower than that of gene coding regions. Hereby, non-conserved regions should not be ignored. To score the conservation of the sequences independently is an alternative way to solve this problem [72].
With the development of high-throughput sequencing, some public experimental data can be incorporated to identify the miRNA-target interaction. For example, paired expression profiles of miRNAs and mRNAs have been used to identify functional miRNA-target [76]. Ago binding sequences and nuclear/cytoplasmic localization data from previous deep sequencing experiment have been incorporated for miRNA-promoter interaction prediction [77].
Public software tools
MicroPIR2 (http://www4a.biotec.or.th/micropir2/) predicts miRNA target of mouse and human promoter region with related genomic and experimental information. The nuclear/cytoplasmic localization and data obtained from some studies and experimentally verified binding sites of Ago proteins are also incorporated into the database [77].
MiRwalk2 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/) provides predicted and validated information of miRNA-target interaction. The binding sites of miRNA can be set to be within all regions of a gene (promoter, 5’-UTR, CDS and 3’-UTR). Results of other 12 miRNA-target prediction programs are also combined and analyzed in miRwalk2 [78, 79].
Trident (http://trident.stjude.org) is a computational algorithm to identify Hoogsteen and reverse Hoogsteen interactions between single-stranded oligonucleotides (microRNA) and double-stranded oligonucleotides (double-stranded DNA). The thermodynamic binding score and heuristic score are measured to evaluate the interaction of miRNA and DNA. Higher heuristic score and lower thermodynamic energy indicate a stronger interaction [63].
Other traditional miRNA target prediction software tools or algorithms can also be used for nuclear miRNA analysis. In order to prevent false positive results, some miRNA target prediction algorithms require conservation of seed sequence and limit the target site in the 3' UTR of mRNA. However, the nuclear miRNAs-targeted promoters are always poorly conserved. There are useful software tools with less limitations or more optional parameters such as TargetS (http://liubioinfolab.org/targetS/mirna.html) [72], RegRNA 2.0 (http://regrna2.mbc.nctu.edu.tw/index.html) [80], and some downloadable applications such as miRanda [81] and RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid) [74].
MicroRNA nuclear functions in immunity
Granulopoiesis
Some microRNAs have been identified as key modulators of granulopoiesis through detection of the miRNA expression profiles at different stages of hematopoietic differentiation [32, 82, 83]. Several miRNAs predominantly localize in the nucleus in murine myeloid cells, such as miR-223, miR-706, miR-690, miR-709 and miR-467a* [32, 84]. In the cytoplasm of myeloid progenitor cells, miR-223 targets transcription factors MEF2C and NF1A, resulting in increased granulopoiesis. However, in the nucleus, miR-223 also targets the NF1A promoter, recruiting the polycomb repressive complex and increasing the DNA methylation and repressive histone markers [84]. Thus, miR-223 could be an essential determination factor of hematopoietic proliferation and differentiation.
Cytokine
Nuclear microRNAs have the ability to regulate the activation of immune cells via expression regulation of cellular cytokines. For example, let7i binds to the TATA-box and activates the transcription of interleukin-2 (IL-2) in CD4+ T-lymphocytes [60], which is an early landmark critical in the activation of CD4+ T cells [85]. In prostate carcinoma cells, miR-205 activates IL-24 and IL-32 by targeting their promoters. IL-24 and IL-32 are members of cytokine family both serving as tumor suppressor [86]. IL-24 is also known as melanoma differentiation-associated-7 because of its tumor suppression function. It is mainly expressed and functions in non-hematopoietic tissues as an inducer of cell death [87]. Moreover, IL-32 is a proinflammatory cytokine, which also functions in cell differentiation, activation of NK and NKT cells, maturation and activation of immature DCs and infection control, etc. [88]. Thus, nuclear microRNA may play an important role in the pathway of immune response and signal transduction by targeting the promoter of cytokine genes.
Inflammation
Two adjacent inflammatory genes, cyclooxygenase-2 (COX-2) and phospholipase A2 (PLA2G4A) are both activated by miR-589. The targets of miR-589 are located in the promoter of COX-2, and gene looping facilitates the interaction between promoters of COX-2 and PLA2G4A. PLA2G4A encodes an enzyme which catalyzes hydrolysis of membrane phospholipids to release arachidonic acid. COX-2 catalyzes the conversion of arachidonic acid to prostaglandins, an inflammatory factor which plays an important role in vasodilation [60].
Asthma
CD44, a transmembrane glycoprotein, plays a vital role in a variety of immune processes including lymphocyte activation, cell proliferation and assembly of inflammatory cells. Recently, some studies [89, 90] demonstrated that CD44 is associated with airway inflammation, particularly asthma, where Li et al. [91] found high expression level of miR-31 in asthma patients. Further study revealed that miR-31 can directly bind to the promoter of CD44 gene and upregulate its expression. Overexpressed CD44 could further induce the expression of asthma-associated molecules, including IL-6, IL-8 and intercellular adhesion molecule 1 (ICAM), which promote progression of asthma.
MicroRNA nuclear functions in cancer
Cancer progression is a metabolically dynamic process that involves tumor initiation, promotion, progression, angiogenesis and metastasis. In a classic cytoplasmic manner, numerous studies illustrate the significant role miRNAs play in cancer. For example, In glioma, miR-200c represses tumor growth and metastasis via interaction with moesin (MSN) mRNA [92]. MiR-4260 serves as a oncopromoter in the colorectal cancer by targeting tumor suppressor colorectal mutant cancer protein (MCC) and SMAD4 [93].
Additionally, nuclear miRNAs have been reported to be involved in the transcriptional regulation of a variety of tumor promoter/repressor genes or genes that are cancer-related by acting upon the promoter of the respective gene loci. Intriguingly, other acting mechanisms of miRNA have also been reported [94]. Over the recent years, a burgeoning literature has been focused on nuclear miRNAs and cancer. Despite ongoing challenges, sizable results have been achieved. Here we attempt to review the current progress of this research endeavor (Table 3).
Tumor initiation, self-sustenance and apoptosis
It is of general consensus that cancer initiation and progression results from aberrant control and integration of growth, differentiation and apoptosis regulatory signals, which could lead to immortalized cell groups capable of self-sustenance and auto-renewal [95]. Apoptosis is an orchestrated and ordered cellular process in which cells experience programmed cell death under physiological and pathological conditions. Defects can occur at any point along apoptotic pathways, leading to malignant transformation of the affected cells, tumor metastasis and drug resistance [96]. Recent researches in nuclear miRNA and transcriptional gene silencing/activation mechanisms have provided insights that could enrich our understanding in tumorigenesis and apoptosis processes.
Studies using chromatin immunoprecipitation and RNA mimicking reveal that miR-423-5p decreases RNA polymerase II occupancy and increases histone H3 lysine 9 dimethylation (H3K9me2) at the progesterone receptor (PR) promoter of human breast cancer cells, indicating a chromatin-level silencing mechanism for the regulation of expression of PR, which mediates endocrinal effects in the development of the mammary gland and breast cancer [97].
MiR-483, an oncogenic intronic miRNA, is reported to bind to the most upstream imprinted insulin-like growth factor 2 (IGF2) gene promoter P2. Ectopic expression of miR-483 induces upregulation of IGF2 expression, as well as an increase in tumor cell proliferation, migration, invasion, and tumor colony formation [98].
Alpha-methylacyl-CoA racemase (AMACR) is highly overexpressed in prostate cancer (PCa) and its transcriptional regulators include various transcription factors and CTNNB1/β-catenin. Studies conducted in vitro in PCacells by Erdmann K et al. [99] revealed that miR-138 indirectly up-regulates AMACR via transcriptional induction of CTNNB1.
Three miRNAs (miR-370, miR-1180 and miR-1236) induce nuclear p21 expression through p21-promoter binding. The expression levels of three miRNAs decrease in bladder cancer tissues compared to healthy controls. Meanwhile, expression of these miRNAs positively correlate with p21 mRNA expression. Overexpression of these three miRNAs inhibits the proliferation of bladder cancer cells mainly by regulating p21 [100].
MiR-211 is a pro-survival microRNA that down-regulates the pro-apoptotic transcription factor C/EBP-homologous protein (CHOP) in a stress and PERK-dependent manner. This permits the cell to prevent premature accumulation of CHOP and rebuild homeostasis prior to apoptotic activation [101]. Studies using functional proteomics demonstrated a RNA-activation function of miR-124 resulting in direct induction of p27 protein expression by binding to and inducing transcription on the p27 promoter region, leading to a subsequent G1 arrest. Ensuing in vivo studies utilizing a xenograft model demonstrated that nanoparticle-mediated delivery of miR-124 could reduce tumor growth and sensitize cells to etoposide to increase apoptosis [102]. Similarly, miR-6734 was found to inhibit the growth of colon cancer cells by up-regulating p21 gene expression and subsequent induction of cell cycle arrest and apoptosis [103]. The binding site of miR-877-3p was also found on the promoter site of tumor suppressor gene p16. P16 exerts a similar function on inhibiting the proliferation and tumorigenesis of bladder cancer through induction of G1 arrest [104]. P16/INK4a is hypothesized to modulate EPCs senescence with telomerase [105].
Overall, nuclear miRNAs, which mainly act upon the promoter regions, are observed to affect the expression profile of oncogenes, tumor suppressors or other cancer-related genes in the cancer initiation process.
Metastasis and angiogenesis
Tumor metastasis involves development of genetic and epigenetic alterations in malignant cells, which could result in global dissemination of cancer cells and life-threatening conditions in patients. Angiogenesis in a key step in the metastatic cascade and provides the primary tumor the main route for transport through the vasculature [106]. A number of metastasis regulators have been discovered in recent years that are targets of miRNA endonuclear mechanisms, including matrix metalloproteinase (MMP) [107,108,109], protocadherin 9 (PCDH9) [110], E-cadherin [2], cold shock domain containing C2 (CSDC2) [2], etc., many of which are involved in the regulation of extracellular matrix (ECM) integrity or effectors of the epithelial–mesenchymal transition.
E-cadherin belongs to cadherin superfamily and functions as adhesion regulator between cells. This gene is considered a tumor repression gene, the loss of which is observed in many malignant tumors. MiR-373 induces expression of E-cadherin and CSDC2 through promoter binding [2]. MiR-205 increases the expression of tumor suppressor genes IL24 and IL32 in prostate cancer. Transfection of miR-205 leads to decrease of cell growth, metastasis and invasiveness of prostate cancer cells [86].
Matrix metalloproteinase 14 (MMP-14) is a membrane-anchored MMP that promotes migration and invasiveness in various tumors. Both miR-337-3p and miR-584-5p inhibit the transcription of MMP-14 in human neuroblastoma. Overexpression of both miRNAs leads to decrease of growth, metastasis and angiogenesis of human neuroblastoma in vitro and in vivo [108, 111]. In gastric cancer, miR-337-3p inhibit myeloid zinc finger 1 (MZF1) induced transcriptional activation of MMP-14 through recruiting Ago2 and inducing repressive chromatin remodeling. MiR-337-3p attenuates growth, migration, and angiogenesis of gastric cancer cells in vitro and in vivo [109].
MiR-558 can bind to the promoter of Heparanase (HPSE) and enhance its expression activity in an Ago-1-dependent manner. Heparanase is an endoglycosidase which degrades polymeric heparan sulfate molecules and is associated with migration and invasiveness of tumor, like in choriocarcinoma [112]. Consistently, knockdown and over-expression of miR-558 indicate its positive function on tumorigenesis and aggressiveness in neuroblastoma cells [107].
Protocadherin 9 (PCDH9), a member of cadherin superfamily, facilitates cell-cell adhesion and is downregulated in glioma. MiR-215-5p binds to both promoter and 3’ UTR of PCDH9 and inhibits the expression of PCDH9 in glioma [110].
Metastasis Associated Lung Adenocarcinoma Transcript 1 (MALAT-1), a nuclear long non-coding RNA, plays an important role in the metastasis in non-small-cell lung carcinoma (NSCLC). In previous studies, miR-9 post-transcriptionally regulate MALAT1 in an Ago2-dependent manner in the nucleus [48].
Application of RNAa in cancer therapeutics
In addition to RNA activation (RNAa) triggered by endogenous miRNAs as previously described [2, 86, 100, 102, 107, 113], RNAa has also been exploited and applied in researches of cancer therapeutics and tumorigenesis. Tumorigenesis is a complex process that often entails underexpression of a variety of tumor suppressor genes involved in multiple signal transduction pathways. RNAa, which is highly specific and poses far less danger to genome integrity than viral vectors, could be utilized to upregulate a cohort of tumor suppressors and thus relieve tumor progression. In vivo and in vitro studies have been conducted with effective tumor inhibitory outcomes [114,115,116,117,118,119].
P21 (also referred to as cyclin-dependent kinase inhibitor 1 or CDK-interacting protein 1) is a tumor suppressor and an important cell cycle regulator which relays the upstream effect of p53 gene and inhibits cyclin-dependent kinase (CDK). Overexpression of p21 results in inhibition of cell-cycle progression and G1 arrest, thus could potentially repress tumor progress [120]. In vivo and in vitro studies of p21 RNAa have been conducted in researches where inhibition of proliferation in prostate, lung, hepatocellular, pancreatic and bladder cancer cells was observed [118, 121,122,123,124]. In another study, hepatocellular carcinoma cells were transfected with dsRNA by liposomes targeting the Wilms’ tumor 1 (WT1) gene promoter, which upregulated WT1, a potent tumor suppressor, and resulted in increased apoptosis of malignant cells [125]. The selected small activating RNA (saRNA) also inhibited the growth, invasion and migration of GC cells by specially reactivating vezatin (VEZT), which resulted in an obvious decrease in the proliferative, invasive and migratory abilities of cancer cells [126]. Moreover, RNAa has been used to promote E-cadherin expression and repress tumor invasion and metastasis in vivo and in vitro in breast, prostate and bladder cancer [116, 127, 128].
Additionally, RNAa has been studied in sensitizing tumor cells to chemotherapy. Innate or acquired resistance to chemotherapy is a formidable challenge that confronts oncologists. Recently, cisplatin nanoparticles co-loaded with miR-375 was developed as a promising treatment for the hepatocellular carcinoma (HCC) based on classic theory of interaction between miRNA and mRNA [129]. Moreover, in one study, saRNA targeting to p21 was transfected in lung cancer cells that are simultaneously treated with cisplatin. Both in vitro and in vivo experimentations showed transfected groups with enhanced chemosensitivity to cisplatin [118], indicating that saRNA based approaches could be potentially applied to relieve chemotherapy resistance under clinical settings.
Conclusion
Since its discovery in 1993, miRNAs had been assumed as post-transcriptional gene regulators that function by base-pairing with mRNAs to inhibit the translation process. However, in more recent years, it has been demonstrated that miRNA could also play a regulatory role in the nucleus [2, 130]. Meanwhile, shuttling mediators of key proteins in the microRNA pathway have also been found. There is some evidence pointing out that miRNA could direct traditional RISC-mediated post-transcriptional gene silencing (PTGS) activities endonuclearly. MiRNA mainly targets specific nucleotide sequences in the promoter region, recruiting epigenetic remodelers and altering gene expression profiles. MiRNA is also shown to serve as enhancer triggers [66] in the nucleus. Disturbances of nuclear miRNA activities could be implicated in a number of diseases and disorders as well as normal physiological processes in immunity and tumor [60, 88, 98].
Based on different targeting prediction principles, many algorithms have been designed. However, because of the deficiency in sensitivity and specificity, no program is proven superior to the others. Results from 12 different miRNA-target prediction programs are integrated into the miRwalk2 database. This database also provides validated target information by an automated text-mining search in the titles/abstracts of the PubMed articles. As research continues to reveal miRNA regulatory mechanisms, the algorithms will become more sensitive and accurate [78, 79, 131].
However, some early results are not repeatable and even conflict with later results. One example is the hexanucleotide element for miR-29b nuclear entrance [33]. MiR-29a without this element will be retained in cytoplasm. However, enrichment of miR-29b is not observed in other cell lines [28]. One explanation is that localization varies among different cell lines. Another explanation is error due to technique. Purity of nuclear abstraction is hard to guarantee. Recently, a protocol for nucleus-cytoplasm division was developed to solve this problem [132].
Nowadays, miRNA therapy and related researches are in a trend to becoming more and more common, but most of them are based on cytoplasmic functions rather than nuclear functions [133]. Accumulating evidence urge us to consider the yet not-too-familiar effect miRNA exerts on the transcription level. As our understanding of its function and dysregulation deepens, nuclear miRNA promises to have applications in a number of clinical settings in the future [4].
References
Lee RC, Feinbaum RL, Ambros V. The C. Elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54.
Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105:1608.
Morris KV, Chan SWL, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–92.
Ross JP, Kassir Z. The varied roles of nuclear argonaute-small RNA complexes and avenues for therapy. Mol Ther-Nucleic Acids. 2014;3:e203.
Schraivogel D, Meister G. Import routes and nuclear functions of Argonaute and other small RNA-silencing proteins. Trends Biochem Sci. 2014;39:420–31.
Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–55.
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33.
Kwon SC, Nguyen TA, Choi YG, Jo MH, Hohng S, Kim VN, Woo JS. Structure of human DROSHA. Cell. 2016;164:81–90.
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9.
Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–6.
Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011;475:201–5.
Wilson RC, Tambe A, Kidwell MA, Noland CL, Schneider CP, Doudna JA. Dicer-TRBP complex formation ensures accurate mammalian microRNA biogenesis. Mol Cell. 2015;57:397–407.
Kwak PB, Tomari Y. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat Struct Mol Biol. 2012;19:145–51.
Jia J, Shi Y, Chen L, Lai W, Yan B, Jiang Y, Xiao D, Xi S, Cao Y, Liu S. Decrease in lymphoid specific Helicase and 5-hydroxymethylcytosine is associated with metastasis and genome instability. Theranostics. 2017; https://doi.org/10.7150/thno.21389.
Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105.
Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460-71.
Subramanian S Ajay, Brian D Athey, Inhan Lee. Unified translation repression mechanism for microRNAs and upstream AUGs. BMC Genomics. 2010;11(1):155.
Yvonne Tay, Jinqiu Zhang, Andrew M. Thomson, Bing Lim, Isidore Rigoutsos. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455(7216):1124-1128
Weinberg MS, Morris KV. Transcriptional gene silencing in humans. Nucleic Acids Res. 2016;44:6505–17.
Catalanotto C, Cogoni C, Zardo G. MicroRNA in control of gene expression: an overview of nuclear functions. Int J Mol Sci. 2016;17:1712.
Kalantari R, Chiang C-M, Corey DR. Regulation of mammalian transcription and splicing by nuclear RNAi. Nucleic Acids Res. 2015;44:524–37.
Rasko JE, Wong JJ-L. Nuclear microRNAs in normal hemopoiesis and cancer. J Hematol Oncol. 2017;10:8.
Huang, V. Endogenous miRNAa: miRNA-Mediated Gene Upregulation. RNA Activation. Singapore: Springer; 2017. p. 65–79.
Ramchandran, Ramani, and Pradeep Chaluvally-Raghavan. miRNA-mediated RNA activation in mammalian cells. RNA Activation. Singapore: Springer; 2017. p. 81–89.
Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–97.
Politz JCR, Hogan EM, Pederson T. MicroRNAs with a nucleolar location. RNA. 2009;15:1705–15.
Liao J-Y, Ma L-M, Guo Y-H, Zhang Y-C, Zhou H, Shao P, Chen Y-Q, Qu L-H. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS One. 2010;5:e10563.
Park CW, Zeng Y, Zhang X, Subramanian S, Steer CJ. Mature microRNAs identified in highly purified nuclei from HCT116 colon cancer cells. RNA Biol. 2010;7:606–14.
Khudayberdiev SA, Zampa F, Rajman M, Schratt G. A comprehensive characterization of the nuclear microRNA repertoire of post-mitotic neurons. Front Mol Neurosci. 2013; https://doi.org/10.3389/fnmol.2013.00043.
Li ZF, Liang YM, Lau PN, Shen W, Wang DK, Cheung WT, Xue CJ, Poon LM, Lam YW. Dynamic localisation of mature microRNAs in human nucleoli is influenced by exogenous genetic materials. PLoS One. 2013;8:e70869.
B. Chen, B. Zhang, H. Luo, J. Yuan, G. Skogerbø, R. Chen. Distinct microRNA subcellular size and expression patterns in human cancer cells. Int J Cell Biol. 2012;2012:672462.
Wong JJ, Ritchie W, Gao D, Lau KA, Gonzalez M, Choudhary A, Taft RJ, Rasko JE, Holst J. Identification of nuclear-enriched miRNAs during mouse granulopoiesis. J Hematol Oncol. 2014;7:42.
Hwang H-W, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100.
RNALocate: a resource for RNA subcellular localizations. Nucleic Acids Research. 2017.
Robb GB, Brown KM, Khurana J, Rana TM. Specific and potent RNAi in the nucleus of human cells. Nat Struct Mol Biol. 2005;12:133–7.
Rüdel S, Flatley A, Weinmann L, Kremmer E, Meister G. A multifunctional human Argonaute2-specific monoclonal antibody. RNA. 2008;14:1244–53.
Janowski BA, Huffman KE, Schwartz JC, Ram R, Nordsell R, Shames DS, Minna JD, Corey DR. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat Struct Mol Biol. 2006;13:787–92.
Hicks JA, Li L, Matsui M, Chu Y, Volkov O, Johnson KC, Corey DR. Human GW182 Paralogs are the central organizers for RNA-mediated control of transcription. Cell Rep. 2017;20:1543–52.
Wei Y, Li L, Wang D, Zhang C-Y, Zen K. Importin 8 regulates the transport of mature microRNAs into the cell nucleus. J Biol Chem. 2014;289:10270–5.
Weinmann L, Hock J, Ivacevic T, Ohrt T, Mutze J, Schwille P, Kremmer E, Benes V, Urlaub H, Meister G. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell. 2009;136:496–507.
Sharma NR, Wang X, Majerciak V, Ajiro M, Kruhlak M, Meyers C, Zheng ZM. Cell type- and tissue context-dependent nuclear distribution of human Ago2. J Biol Chem. 2016;291:2302–9.
Nishi K, Nishi A, Nagasawa T, Ui-Tei K. Human TNRC6A is an Argonaute-navigator protein for microRNA-mediated gene silencing in the nucleus. RNA. 2013;19:17–35.
D. Schraivogel, S.G. Schindler, J. Danner, E. Kremmer, J. Pfaff, S. Hannus, R. Depping, G. Meister. Importin-β facilitates nuclear import of human GW proteins and balances cytoplasmic gene silencing protein levels, Nucleic Acids Res. 2015;43:7447-7461.
Chaston JJ, Stewart AG, Christie M. Structural characterisation of TNRC6A nuclear localisation signal in complex with importin-alpha. PLoS One. 2017;12:e0183587.
Ohrt T, Mütze J, Staroske W, Weinmann L, Höck J, Crell K, Meister G, Schwille P. Fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy reveal the cytoplasmic origination of loaded nuclear RISC in vivo in human cells. Nucleic Acids Res. 2008;36:6439–49.
Kalantari R, Hicks JA, Li L, Gagnon KT, Sridhara V, Lemoff A, Mirzaei H, Corey DR. Stable association of RNAi machinery is conserved between the cytoplasm and nucleus of human cells. RNA. 2016;22:1085–98.
Castanotto D, Lingeman R, Riggs AD, Rossi JJ. CRM1 mediates nuclear-cytoplasmic shuttling of mature microRNAs. Proc Natl Acad Sci. 2009;106:21655–9.
Leucci E, Patella F, Waage J, Holmstrøm K, Lindow M, Porse B, Kauppinen S, Lund AH. microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci Rep. 2013;3:2535.
Tang R, Li L, Zhu D, Hou D, Cao T, Gu H, Zhang J, Chen J, Zhang C-Y, Zen K. Mouse miRNA-709 directly regulates miRNA-15a/16-1 biogenesis at the posttranscriptional level in the nucleus: evidence for a microRNA hierarchy system. Cell Res. 2012;22:504.
Li, L-C. Small RNA-Guided Transcriptional Gene Activation (RNAa) in Mammalian Cells. RNA Activation. Singapore: Springer; 2017. p. 1-20.
Li L-C. Chromatin remodeling by the small RNA machinery in mammalian cells. Epigenetics. 2014;9:45–52.
Li L-C, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci. 2006;103:17337–42.
Meng X, Jiang Q, Chang N, Wang X, Liu C, Xiong J, Cao H, Liang Z. Small activating RNA binds to the genomic target site in a seed-region-dependent manner. Nucleic Acids Res. 2016;44:2274–82.
He X, Yan B, Liu S, Jia J, Lai W, Xin X, Tang C-e, Luo D, Tan T, Jiang Y. Chromatin remodeling factor LSH drives cancer progression by suppressing the activity of fumarate hydratase. Cancer Res. 2016;76:5743–55.
Zheng L, Chen Y, Ye L, Jiao W, Song H, Mei H, Li D, Yang F, Li H, Huang K, Tong Q. miRNA-584-3p inhibits gastric cancer progression by repressing Yin Yang 1- facilitated MMP-14 expression. Scientific reports. 2017;7:8967
Portnoy V, Lin SHS, Li KH, Burlingame A, Hu Z-H, Li H, Li L-C. saRNA-guided Ago2 targets the RITA complex to promoters to stimulate transcription. Cell Res. 2016;26:320.
Ting AH, Schuebel KE, Herman JG, Baylin SB. Short dsRNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat Genet. 2005;37:906.
Huang V, Place RF, Portnoy V, Wang J, Qi Z, Jia Z, Yu A, Shuman M, Yu J, Li L-C. Upregulation of Cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res. 2011;40:1695–707.
Schwartz JC, Younger ST, Nguyen N-B, Hardy DB, Monia BP, Corey DR, Janowski BA. Antisense transcripts are targets for activating small RNAs. Nat Struct Mol Biol. 2008;15:842–8.
Matsui M, Chu Y, Zhang H, Gagnon KT, Shaikh S, Kuchimanchi S, Manoharan M, Corey DR, Janowski BA. Promoter RNA links transcriptional regulation of inflammatory pathway genes. Nucleic Acids Res. 2013;41:10086–109.
Pisignano G, Napoli S, Magistri M, Mapelli SN, Pastori C, Di Marco S, Civenni G, Albino D, Enriquez C, Allegrini S. A promoter-proximal transcript targeted by genetic polymorphism controls E-cadherin silencing in human cancers. Nat Commun. 2017;8:15622
Morris KV, Santoso S, Turner A-M, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:e1000258.
Bethany A Janowski, Kenneth E Huffman, Jacob C Schwartz, Rosalyn Ram, Daniel Hardy, David S Shames, John D Minna, David R Corey. Inhibiting gene expression at transcription start sites in chromosomal DNA with antigene RNAs. Nature Chemical Biology. 2005;1(4):216-222
Yue X, Schwartz JC, Chu Y, Younger ST, Gagnon KT, Elbashir S, Janowski BA, Corey DR. Transcriptional regulation by small RNAs at sequences downstream from 3′ gene termini. Nat Chem Biol. 2010;6:621–9.
Zhang Y, Fan M, Zhang X, Huang F, Wu K, Zhang J, Liu J, Huang Z, Luo H, Tao L. Cellular microRNAs up-regulate transcription via interaction with promoter TATA-box motifs. RNA. 2014;20:1878–89.
Li Z, Lan X, Han R, Wang J, Huang Y, Sun J, Guo W, Chen H. miR-2478 inhibits TGFβ1 expression by targeting the transcriptional activation region downstream of the TGFβ1 promoter in dairy goats. Sci Rep. 2017;7:42627.
Zhang Y, Zhang H. RNAa Induced by TATA Box-Targeting MicroRNAs. RNA Activation. Singapore: Springer; 2017. p. 91-111.
Paugh SW, Coss DR, Bao J, Laudermilk LT, Grace CR, Ferreira AM, Waddell MB, Ridout G, Naeve D, Leuze M. MicroRNAs form triplexes with double stranded DNA at sequence-specific binding sites; a eukaryotic mechanism via which microRNAs could directly alter gene expression. PLoS Comput Biol. 2016;12:e1004744.
Toscano-Garibay JD, Aquino-Jarquin G. Transcriptional regulation mechanism mediated by miRNA–DNA• DNA triplex structure stabilized by Argonaute. Bioch Biophys Acta, Gene Regul Mech. 2014;1839:1079–83.
Kanak M, Alseiari M, Balasubramanian P, Addanki K, Aggarwal M, Noorali S, Kalsum A, Mahalingam K, Pace G, Panasik N. Triplex-forming MicroRNAs form stable complexes with HIV-1 provirus and inhibit its replication. Appl Immunohistochem Mol Morphol. 2010;18:532–45.
Xiao M, Li J, Li W, Wang Y, Wu F, Xi Y, Zhang L, Ding C, Luo H, Li Y. MicroRNAs activate gene transcription epigenetically as an enhancer trigger. RNA Biol. 2016:00–0.
Xu W, San Lucas A, Wang Z, Liu Y. Identifying microRNA targets in different gene regions. BMC Bioinf. 2014;15:S4.
Younger ST, Corey DR. Transcriptional gene silencing in mammalian cells by miRNA mimics that target gene promoters. Nucleic Acids Res. 2011;39:5682–91.
Rehmsmeier M, Steffen P, Höchsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–17.
De Witte D, Van de Velde J, Decap D, Van Bel M, Audenaert P, Demeester P, Dhoedt B, Vandepoele K, Fostier J. BLSSpeller: exhaustive comparative discovery of conserved cis-regulatory elements. Bioinformatics. 2015;31:3758–66.
Huang JC, Babak T, Corson TW, Chua G, Khan S, Gallie BL, Hughes TR, Blencowe BJ, Frey BJ, Morris QD. Using expression profiling data to identify human microRNA targets. Nat Methods. 2007;4:1045–9.
Piriyapongsa J, Bootchai C, Ngamphiw C, Tongsima S. microPIR2: a comprehensive database for human–mouse comparative study of microRNA–promoter interactions. Database. 2014;2014:bau115.
Dweep H, Gretz N. miRWalk2. 0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12:697.
Parveen A, Gretz N, Dweep H. Obtaining miRNA-Target Interaction Information from miRWalk2. 0. Cur Prot Bioinformatics. 2016;55:12.15.1-12.15.27.
Chang T-H, Huang H-Y, Hsu JB-K, Weng S-L, Horng J-T, Huang H-D. An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinf. 2013;14:S4.
John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2:e363.
Larsen MT, Hother C, Häger M, Pedersen CC, Theilgaard-Mönch K, Borregaard N, et al. MicroRNA Profiling in Human Neutrophils during Bone Marrow Granulopoiesis and In Vivo Exudation. PLoS ONE. 2013;8:e58454.
Sun SM, Dijkstra MK, Bijkerk AC, Brooimans RA, Valk PJ, Erkeland SJ, Löwenberg B, Jongen-Lavrencic M. Transition of highly specific microRNA expression patterns in association with discrete maturation stages of human granulopoiesis. Br J Haematol. 2011;155:395–8.
Zardo G, Ciolfi A, Vian L, Starnes LM, Billi M, Racanicchi S, Maresca C, Fazi F, Travaglini L, Noguera N. Polycombs and microRNA-223 regulate human granulopoiesis by transcriptional control of target gene expression. Blood. 2012;119:4034–46.
Sojka DK, Bruniquel D, Schwartz RH, Singh NJ. IL-2 secretion by CD4+ T cells in vivo is rapid, transient, and influenced by TCR-specific competition. J Immunol. 2004;172:6136–43.
Majid S, Dar AA, Saini S, Yamamura S, Hirata H, Tanaka Y, Deng G, Dahiya R. MicroRNA-205–directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer. 2010;116:5637–49.
Kreis S, Philippidou D, Margue C, Behrmann I. IL-24: a classic cytokine and/or a potential cure for cancer? J Cell Mol Med. 2008;12:2505–10.
Ribeiro-Dias F, Gomes RS, de Lima Silva LL, dos Santos JC, Joosten LA. Interleukin 32: a novel player in the control of infectious diseases. J Leukoc Biol. 2017;101:39–52.
Kumar S, Lanckacker E, Dentener M, Bracke K, Provoost S, De Grove K, Brusselle G, Wouters E, Maes T, Joos G. Aggravation of allergic airway inflammation by cigarette smoke in mice is CD44-dependent. PLoS One. 2016;11:e0151113.
Johansson MW. Eosinophil activation status in separate compartments and association with asthma. Front Med. 2017;4:75.
Li L, Hui Y, Xing C, Guo Y, Wang Q, Shu J, Qian J, Zhou G. MicroRNA-31 affects the expression of asthma-related cytokines via regulation of CD44. Int. J Clin Exp Med. 2016;9:11.
Qin Y, Chen W, Liu B, Zhou L, Deng L, Niu W, Bao D, Cheng C, Li D, Liu S. MiR-200c inhibits the tumor progression of Glioma via targeting Moesin. Theranostics. 2017;7:1663.
Xiao J, Lv D, Zhou J, Bei Y, Chen T, Hu M, Zhou Q, Fu S, Huang Q. Therapeutic inhibition of miR-4260 suppresses colorectal cancer via targeting MCC and SMAD4. Theranostics. 2017;7:1901.
Cui C, Yu J, Huang S, Zhu H, Huang Z. Transcriptional regulation of gene expression by microRNAs as endogenous decoys of transcription factors. Cell Physiol Biochem. 2014;33:1698–714.
Scott RE, Wille JJ Jr, Wier ML. Mechanisms for the initiation and promotion of carcinogenesis: a review and a new concept. Mayo Clin Proc. 1984;59:107–17.
Hassan M, Watari H, AbuAlmaaty A, Ohba Y, Sakuragi N. Apoptosis and molecular targeting therapy in cancer. Biomed Res Int. 2014;2014:150845.
Conneely OM, Jericevic BM, Lydon JP. Progesterone receptors in mammary gland development and tumorigenesis. J Mammary Gland Biol Neoplasia. 2003;8:205–14.
Zhang Y, Hu JF, Wang H, Cui J, Gao S, Hoffman AR, Li W. CRISPR Cas9-guided chromatin immunoprecipitation identifies miR483 as an epigenetic modulator of IGF2 imprinting in tumors. Oncotarget. 2017;8:34177–90.
Erdmann K, Kaulke K, Rieger C, Wirth MP, Fuessel S. Induction of alpha-methylacyl-CoA racemase by miR-138 via up-regulation of beta-catenin in prostate cancer cells. Journal of cancer research and clinical oncology. 2017;143:2201-2210.
Wang C, Chen Z, Ge Q, Hu J, Li F, Hu J, Xu H, Ye Z, Li LC. Up-regulation of p21(WAF1/CIP1) by miRNAs and its implications in bladder cancer cells. FEBS Lett. 2014;588:4654–64.
Chitnis NS, Pytel D, Bobrovnikova-Marjon E, Pant D, Zheng H, Maas NL, Frederick B, Kushner JA, Chodosh LA, Koumenis C, Fuchs SY, Diehl JA. miR-211 is a prosurvival microRNA that regulates chop expression in a PERK-dependent manner. Mol Cell. 2012;48:353–64.
Seviour EG, Sehgal V, Lu Y, Luo Z, Moss T, Zhang F, Hill SM, Liu W, Maiti SN, Cooper L, Azencot R, Lopez-Berestein G, Rodriguez-Aguayo C, Roopaimoole R, Pecot CV, Sood AK, Mukherjee S, Gray JW, Mills GB, Ram PT. Functional proteomics identifies miRNAs to target a p27/Myc/phospho-Rb signature in breast and ovarian cancer. Oncogene. 2016;35:691–701.
Kang MR, Park KH, Yang JO, Lee CW, Oh SJ, Yun J, Lee MY, Han SB, Kang JS. miR-6734 up-regulates p21 gene expression and induces cell cycle arrest and apoptosis in colon cancer cells. PLoS One. 2016;11:e0160961.
Li S, Zhu Y, Liang Z, Wang X, Meng S, Xu X, Xu X, Wu J, Ji A, Hu Z, Lin Y, Chen H, Mao Y, Wang W, Zheng X, Liu B, Xie L. Up-regulation of p16 by miR-877-3p inhibits proliferation of bladder cancer. Oncotarget. 2016;7:51773–83.
Yang DG, Liu L, & Zheng XY. Cyclin-dependent kinase inhibitor p16 ink4a, and telomerase may co-modulate endothelial progenitor cells senescence. Ageing Res Rev. 2008;7(2):137.
Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–92.
Qu H, Zheng L, Pu J, Mei H, Xiang X, Zhao X, Li D, Li S, Mao L, Huang K. miRNA-558 promotes tumorigenesis and aggressiveness of neuroblastoma cells through activating the transcription of heparanase. Hum Mol Genet. 2015;24:2539–51.
Xiang X, Mei H, Zhao X, Pu J, Li D, Qu H, Jiao W, Zhao J, Huang K, Zheng L. miRNA-337-3p suppresses neuroblastoma progression by repressing the transcription of matrix metalloproteinase 14. Oncotarget. 2015;6:22452.
Zheng L, Jiao W, Mei H, Song H, Li D, Xiang X, Chen Y, Yang F, Li H, Huang K. miRNA-337-3p inhibits gastric cancer progression through repressing myeloid zinc finger 1-facilitated expression of matrix metalloproteinase 14. Oncotarget. 2016;7:40314.
Wang C, Chen Q, Li S, Li S, Zhao Z, Gao H, Wang X, Li B, Zhang W, Yuan Y. Dual inhibition of PCDH9 expression by miR-215-5p up-regulation in gliomas. Oncotarget. 2017;8:10287.
Xiang X, Mei H, Qu H, Zhao X, Li D, Song H, Jiao W, Pu J, Huang K, Zheng L. miRNA-584-5p exerts tumor suppressive functions in human neuroblastoma through repressing transcription of matrix metalloproteinase 14. Biochim Biophy Acta (BBA)-Mol Basis Dis. 2015;1852:1743–54.
Jingting C, et al. Heparanase expression correlates with metastatic capability in human choriocarcinoma. Gynecol Oncol. 2007;107(1):22–9.
Liu M, Roth A, Yu M, Morris R, Bersani F, Rivera MN, Lu J, Shioda T, Vasudevan S, Ramaswamy S, Maheswaran S, Diederichs S, Haber DA. The IGF2 intronic miR-483 selectively enhances transcription from IGF2 fetal promoters and enhances tumorigenesis. Genes Dev. 2013;27:2543–8.
Chen R, Wang T, Rao K, Yang J, Zhang S, Wang S, Liu J, Ye Z. Up-regulation of VEGF by small activator RNA in human corpus cavernosum smooth muscle cells. J Sex Med. 2011;8:2773–80.
Dong Z, Dang Y, Chen Y. Small double-stranded RNA mediates the anti-cancer effects of p21WAF1/ClP1 transcriptional activation in a human glioma cell line. Yonsei Med J. 2014;55:324–30.
Junxia W, Ping G, Yuan H, Lijun Z, Jihong R, Fang L, Min L, Xi W, Ting H, Ke D, Huizhong Z. Double strand RNA-guided endogeneous E-cadherin up-regulation induces the apoptosis and inhibits proliferation of breast carcinoma cells in vitro and in vivo. Cancer Sci. 2010;101:1790–6.
Wang J, Li LC. Small RNA and its application in andrology and urology. Transl Androl Urol. 2012;1:33–43.
Wei J, Zhao J, Long M, Han Y, Wang X, Lin F, Ren J, He T, Zhang H. p21WAF1/CIP1 gene transcriptional activation exerts cell growth inhibition and enhances chemosensitivity to cisplatin in lung carcinoma cell. BMC Cancer. 2010;10:632.
Yang K, Shen J, Xie YQ, Lin YW, Qin J, Mao QQ, Zheng XY, Xie LP. Promoter-targeted double-stranded small RNAs activate PAWR gene expression in human cancer cells. Int J Biochem Cell Biol. 2013;45:1338–46.
Hoeferlin LA, Oleinik NV, Krupenko NI, Krupenko SA. Activation of p21-dependent G1/G2 arrest in the absence of DNA damage as an Antiapoptotic response to metabolic stress. Genes Cancer. 2011;2:889–99.
Place RF, Wang J, Noonan EJ, Meyers R, Manoharan M, Charisse K, Duncan R, Huang V, Wang X, Li LC. Formulation of small activating RNA into Lipidoid Nanoparticles inhibits Xenograft prostate tumor growth by inducing p21 expression. Mol Ther Nucleic Acids. 2012;1:e15.
Kosaka M, Kang MR, Yang G, Li LC. Targeted p21WAF1/CIP1 activation by RNAa inhibits hepatocellular carcinoma cells. Nucleic Acid Ther. 2012;22:335–43.
Zhang Z, Wang Z, Liu X, Wang J, Li F, Li C, Shan B. Up-regulation of p21WAF1/CIP1 by small activating RNA inhibits the in vitro and in vivo growth of pancreatic cancer cells. Tumori. 2012;98:804–11.
Yang K, Zheng XY, Qin J, Wang YB, Bai Y, Mao QQ, Wan Q, Wu ZM, Xie LP. Up-regulation of p21WAF1/Cip1 by saRNA induces G1-phase arrest and apoptosis in T24 human bladder cancer cells. Cancer Lett. 2008;265:206-214.
Qin Q, Lin Y-W, Zheng X-Y, Chen H, Mao Q-Q, Yang K, Huang S-J, Zhao Z-Y. RNAa-mediated overexpression of WT1 induces apoptosis in HepG2 cells. World J Surg Oncol. 2012;10:11.
Xie D, Shang L, Peng L, Li L. Up-regulation of VEZT by small activating RNA inhibits the proliferation, invasion and migration of gastric cancer cells. Biochem Biophys Res Commun. 2017;482:542–8.
Mao Q, Li Y, Zheng X, Yang K, Shen H, Qin J, Bai Y, Kong D, Jia X, Xie L. Up-regulation of E-cadherin by small activating RNA inhibits cell invasion and migration in 5637 human bladder cancer cells. Biochem Biophys Res Commun. 2008;375:566–70.
Mao Q, Zheng X, Yang K, Qin J, Bai Y, Jia X, Li Y, Xie L. Suppression of migration and invasion of PC3 prostate cancer cell line via activating E-cadherin expression by small activating RNA. Cancer Investig. 2010;28:1013–8.
Yang T, Zhao P, Rong Z, Li B, Xue H, You J, He C, Li W, He X, Lee RJ. Anti-tumor efficiency of lipid-coated cisplatin nanoparticles co-loaded with microRNA-375. Theranostics. 2016;6:142.
Kim DH, Saetrom PL, Snøve O, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci U S A. 2008;105:16230-16235.
Dweep H, Sticht C, Gretz N. In-silico algorithms for the screening of possible microRNA binding sites and their interactions. Cur Genomics. 2013;14:127–36.
Gagnon KT, Li L, Janowski BA, Corey DR. Analysis of nuclear RNA interference in human cells by subcellular fractionation and Argonaute loading. Nat Protoc. 2014;9:2045.
E. Van Rooij, S. Kauppinen. Development of microRNA therapeutics is coming of age. EMBO Mol Med. 2014;6:851-864.
Taft RJ, Simons C, Nahkuri S, Oey H, Korbie DJ, Mercer TR, Holst J, Ritchie W, Wong JJ, Rasko JE. Nuclear-localized tiny RNAs are associated with transcription initiation and splice sites in metazoans. Nat Struct Mol Biol. 2010;17:1030–4.
Li Z, Lan X, Han R, Wang J, Huang Y, Sun J, Guo W, Chen H. miR-2478 inhibits TGFbeta1 expression by targeting the transcriptional activation region downstream of the TGFbeta1 promoter in dairy goats. Sci Rep. 2017;7:42627.
Tan Y, Zhang B, Wu T, Skogerbo G, Zhu X, Guo X, He S, Chen R. Transcriptional inhibiton of Hoxd4 expression by miRNA-10a in human breast cancer cells. BMC Mol Biol. 2009;10:12.
Xiang X, Mei H, Qu H, Zhao X, Li D, Song H, Jiao W, Pu J, Huang K, Zheng L, Tong Q. miRNA-584-5p exerts tumor suppressive functions in human neuroblastoma through repressing transcription of matrix metalloproteinase 14. Biochim Biophys Acta. 2015;1852:1743–54.
Wang C, Chen Q, Li S, Li S, Zhao Z, Gao H, Wang X, Li B, Zhang W, Yuan Y, Ming L, He H, Tao B, Zhong J. Dual inhibition of PCDH9 expression by miR-215-5p up-regulation in gliomas. Oncotarget. 2017;8:10287–97.
Funding
This work was supported by the National Natural Science Foundation of China [81372427 and 81672787(Y.Tao), 81772927(D. Xiao), the National Basic Research Program of China [2015CB553903 (Y.Tao)], and the Fundamental Research Funds for the Central Universities [201710533337 (H.Liu)].
Author information
Authors and Affiliations
Contributions
YT and DX contributed to study conception and design. HL, CL, QH, ZP drafted the manuscript. HL disigned and drew figures. CL, QH, ZP drew tables. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest. This manuscript has been read and approved by all authors, and is not submitted or under consideration for publication elsewhere.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Liu, H., Lei, C., He, Q. et al. Nuclear functions of mammalian MicroRNAs in gene regulation, immunity and cancer. Mol Cancer 17, 64 (2018). https://doi.org/10.1186/s12943-018-0765-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12943-018-0765-5