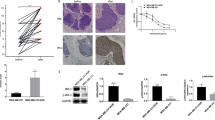
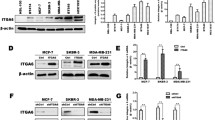
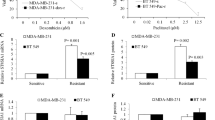
Abstract
Breast cancer is a highly heterogeneous group of human cancer with distinct genetic, biological and clinicopathological features. Triple-negative breast cancer (TNBC) is the most aggressive and metastatic type of breast cancer and associated with poor patient survival. However, the role of UV Radiation Resistance-Associated Gene (UVRAG) in TNBC remains unknown. Here, we report that UVRAG is highly upregulated in all TNBC cells and its knockdown leads to the inhibition of cell proliferation, colony formation and progression of cell cycle, which is associated with and reduced expression of cell cycle related protein expression, including Cyclin A2, B1, D1, cdc2 and cdk6 in TNBC cells. Inhibition of UVRAG also suppressed cell motility, migration and invasion of TNBC cells by inhibition of Integrin β1 and β3 and Src activity. Our findings suggest for the first time that UVRAG expression contributes to proliferation, cell cycle progression, motility/migration and invasion of TNBC cells. Thus, targeting UVRAG could be a potential strategy in breast cancer especially against TNBC.





Similar content being viewed by others
References
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D (2000) Molecular portraits of human breast tumours. Nature 406:747–752. https://doi.org/10.1038/35021093
Rakha EA, El-Sayed ME, Green AR, Lee AH, Robertson JF, Ellis IO (2007) Prognostic markers in triple-negative breast cancer. Cancer 109:25–32. https://doi.org/10.1002/cncr.22381
Audeh MW (2014) Novel treatment strategies in triple-negative breast cancer: specific role of poly(adenosine diphosphate-ribose) polymerase inhibition. Pharmgenomics Pers Med 7:307–316. https://doi.org/10.2147/PGPM.S39765
Dalby KN, Tekedereli I, Lopez-Berestein G, Ozpolat B (2010) Targeting the prodeath and prosurvival functions of autophagy as novel therapeutic strategies in cancer. Autophagy 6:322–329. https://doi.org/10.4161/auto.6.3.11625
Hasima N, Ozpolat B (2014) Regulation of autophagy by polyphenolic compounds as a potential therapeutic strategy for cancer. Cell Death Dis 5:e1509. https://doi.org/10.1038/cddis.2014.467
Ozpolat B, Benbrook DM (2015) Targeting autophagy in cancer management—strategies and developments. Cancer Manag Res 7:291–299. https://doi.org/10.2147/cmar.s34859
Hamurcu Z, Delibasi N, Gecene S, Sener EF, Donmez-Altuntas H, Ozkul Y, Canatan H, Ozpolat B (2018) Targeting LC3 and Beclin-1 autophagy genes suppresses proliferation, survival, migration and invasion by inhibition of cyclin-D1 and uPAR/Integrin beta1/Src signaling in triple negative breast cancer cells. J Cancer Res Clin Oncol 144:415–430. https://doi.org/10.1007/s00432-017-2557-5
Hamurcu Z, Delibaşı N, Nalbantoglu U, Sener EF, Nurdinov N, Tascı B, Taheri S, Özkul Y, Donmez-Altuntas H, Canatan H, Ozpolat B (2019) FOXM1 plays a role in autophagy by transcriptionally regulating Beclin-1 and LC3 genes in human triple-negative breast cancer cells. J Mol Med 97:491–508. https://doi.org/10.1007/s00109-019-01750-8
Hamurcu Z, Ashour A, Kahraman N, Ozpolat B (2016) FOXM1 regulates expression of eukaryotic elongation factor 2 kinase and promotes proliferation, invasion and tumorgenesis of human triple negative breast cancer cells. Oncotarget 7:16619–16635. https://doi.org/10.18632/oncotarget.7672
Tekedereli I, Akar U, Alpay SN, Lopez-Berestein G, Ozpolat B (2019) Autophagy is required to regulate mitochondria renewal, cell attachment, and all-trans-retinoic acid-induced differentiation in NB4 acute promyelocytic leukemia cells. J Environ Pathol Toxicol Oncol 38:13–20. https://doi.org/10.1615/JEnvironPatholToxicolOncol.2018027885
Asik E, Akpinar Y, Caner A, Kahraman N, Guray T, Volkan M, Albarracin C, Pataer A, Arun B, Ozpolat B (2019) EF2-kinase targeted cobalt-ferrite siRNA-nanotherapy suppresses BRCA1-mutated breast cancer. Nanomedicine (London) 14:2315–2338. https://doi.org/10.2217/nnm-2019-0132
Knaevelsrud H, Ahlquist T, Merok MA, Nesbakken A, Stenmark H, Lothe RA, Simonsen A (2010) UVRAG mutations associated with microsatellite unstable colon cancer do not affect autophagy. Autophagy 6:863–870. https://doi.org/10.4161/auto.6.7.13033
Teitz T, Eli D, Penner M, Bakhanashvili M, Naiman T, Timme TL, Wood CM, Moses RE, Canaani D (1990) Expression of the cDNA for the beta subunit of human casein kinase II confers partial UV resistance on xeroderma pigmentosum cells. Mutat Res 236:85–97. https://doi.org/10.1016/0921-8777(90)90036-5
Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mule JJ, Pledger WJ, Wang HG (2007) Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol 9:1142–1151. https://doi.org/10.1038/ncb1634
Yin X, Cao L, Kang R, Yang M, Wang Z, Peng Y, Tan Y, Liu L, Xie M, Zhao Y, Livesey KM, Tang D (2011) UV irradiation resistance-associated gene suppresses apoptosis by interfering with BAX activation. EMBO Rep 12:727–734. https://doi.org/10.1038/embor.2011.79
Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU (2006) Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol 8:688–699. https://doi.org/10.1038/ncb1426
Kim MS, Jeong EG, Ahn CH, Kim SS, Lee SH, Yoo NJ (2008) Frameshift mutation of UVRAG, an autophagy-related gene, in gastric carcinomas with microsatellite instability. Hum Pathol 39:1059–1063. https://doi.org/10.1016/j.humpath.2007.11.013
Kang MR, Kim MS, Oh JE, Kim YR, Song SY, Kim SS, Ahn CH, Yoo NJ, Lee SH (2009) Frameshift mutations of autophagy-related genes ATG2B, ATG5, ATG9B and ATG12 in gastric and colorectal cancers with microsatellite instability. J Pathol 217:702–706. https://doi.org/10.1002/path.2509
Wu T, Li Y, Gong L, Lu JG, Du XL, Zhang WD, He XL, Wang JQ (2012) Multi-step process of human breast carcinogenesis: a role for BRCA1, BECN1, CCND1, PTEN and UVRAG. Mol Med Rep 5:305–312. https://doi.org/10.3892/mmr.2011.634
Hosokawa Y, Arnold A (1998) Mechanism of cyclin D1 (CCND1, PRAD1) overexpression in human cancer cells: analysis of allele-specific expression. Genes Chromosom Cancer 22:66–71. https://doi.org/10.1002/(sici)1098-2264(199805)22:1<66::aid-gcc9>3.0.co;2-5
Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell RG (1995) Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem 270:23589–23597. https://doi.org/10.1074/jbc.270.40.23589
Lee RJ, Albanese C, Stenger RJ, Watanabe G, Inghirami G, Haines GK 3rd, Webster M, Muller WJ, Brugge JS, Davis RJ, Pestell RG (1999) pp60(v-src) induction of cyclin D1 requires collaborative interactions between the extracellular signal-regulated kinase, p38, and Jun kinase pathways. A role for cAMP response element-binding protein and activating transcription factor-2 in pp60(v-src) signaling in breast cancer cells. J Biol Chem 274:7341–7350. https://doi.org/10.1074/jbc.274.11.7341
Lee RJ, Albanese C, Fu M, D’Amico M, Lin B, Watanabe G, Haines GK 3rd, Siegel PM, Hung MC, Yarden Y, Horowitz JM, Muller WJ, Pestell RG (2000) Cyclin D1 is required for transformation by activated Neu and is induced through an E2F-dependent signaling pathway. Mol Cell Biol 20:672–683. https://doi.org/10.1128/mcb.20.2.672-683.2000
Fu M, Wang C, Li Z, Sakamaki T, Pestell RG (2004) Minireview: cyclin D1: normal and abnormal functions. Endocrinology 145:5439–5447. https://doi.org/10.1210/en.2004-0959
Hanai J, Dhanabal M, Karumanchi SA, Albanese C, Waterman M, Chan B, Ramchandran R, Pestell R, Sukhatme VP (2002) Endostatin causes G1 arrest of endothelial cells through inhibition of cyclin D1. J Biol Chem 277:16464–16469. https://doi.org/10.1074/jbc.M112274200
Brown NE, Jeselsohn R, Bihani T, Hu MG, Foltopoulou P, Kuperwasser C, Hinds PW (2012) Cyclin D1 activity regulates autophagy and senescence in the mammary epithelium. Cancer Res 72:6477–6489. https://doi.org/10.1158/0008-5472.CAN-11-4139
Wei H, Wei S, Gan B, Peng X, Zou W, Guan JL (2011) Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev 25:1510–1527. https://doi.org/10.1101/gad.2051011
Koneri K, Goi T, Hirono Y, Katayama K, Yamaguchi A (2007) Beclin 1 gene inhibits tumor growth in colon cancer cell lines. Anticancer Res 27:1453–1457
Choi J, Jung W, Koo JS (2013) Expression of autophagy-related markers beclin-1, light chain 3A, light chain 3B and p62 according to the molecular subtype of breast cancer. Histopathology 62:275–286. https://doi.org/10.1111/his.12002
Lefort S, Joffre C, Kieffer Y, Givel AM, Bourachot B, Zago G, Bieche I, Dubois T, Meseure D, Vincent-Salomon A, Camonis J, Mechta-Grigoriou F (2014) Inhibition of autophagy as a new means of improving chemotherapy efficiency in high-LC3B triple-negative breast cancers. Autophagy 10:2122–2142. https://doi.org/10.4161/15548627.2014.981788
Mathew R, Karantza-Wadsworth V, White E (2007) Role of autophagy in cancer. Nat Rev Cancer 7:961–967. https://doi.org/10.1038/nrc2254
Lazova R, Camp RL, Klump V, Siddiqui SF, Amaravadi RK, Pawelek JM (2012) Punctate LC3B expression is a common feature of solid tumors and associated with proliferation, metastasis, and poor outcome. Clin Cancer Res 18:370–379. https://doi.org/10.1158/1078-0432.CCR-11-1282
Yoshioka A, Miyata H, Doki Y, Yamasaki M, Sohma I, Gotoh K, Takiguchi S, Fujiwara Y, Uchiyama Y, Monden M (2008) LC3, an autophagosome marker, is highly expressed in gastrointestinal cancers. Int J Oncol 33:461–468
Cook KL, Shajahan AN, Clarke R (2011) Autophagy and endocrine resistance in breast cancer. Expert Rev Anticancer Ther 11:1283–1294. https://doi.org/10.1586/era.11.111
Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B (2005) Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122:927–939. https://doi.org/10.1016/j.cell.2005.07.002
Akar U, Chaves-Reyez A, Barria M, Tari A, Sanguino A, Kondo Y, Kondo S, Arun B, Lopez-Berestein G, Ozpolat B (2008) Silencing of Bcl-2 expression by small interfering RNA induces autophagic cell death in MCF-7 breast cancer cells. Autophagy 4:669–679. https://doi.org/10.4161/auto.6083
Cary LA, Guan JL (1999) Focal adhesion kinase in integrin-mediated signaling. Front Biosci 4:D102–D113. https://doi.org/10.2741/cary
Desgrosellier JS, Cheresh DA (2010) Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer 10:9–22. https://doi.org/10.1038/nrc2748
Klahan S, Huang WC, Chang CM, Wong HS, Huang CC, Wu MS, Lin YC, Lu HF, Hou MF, Chang WC (2016) Gene expression profiling combined with functional analysis identify integrin beta1 (ITGB1) as a potential prognosis biomarker in triple negative breast cancer. Pharmacol Res 104:31–37. https://doi.org/10.1016/j.phrs.2015.12.004
Lahlou H, Muller WJ (2011) beta1-integrins signaling and mammary tumor progression in transgenic mouse models: implications for human breast cancer. Breast Cancer Res 13:229. https://doi.org/10.1186/bcr2905
Miranti CK, Brugge JS (2002) Sensing the environment: a historical perspective on integrin signal transduction. Nat Cell Biol 4:E83–E90. https://doi.org/10.1038/ncb0402-e83
Ahn JH, Lee M (2011) Suppression of autophagy sensitizes multidrug resistant cells towards Src tyrosine kinase specific inhibitor PP2. Cancer Lett 310:188–197. https://doi.org/10.1016/j.canlet.2011.06.034
Tuloup-Minguez V, Hamai A, Greffard A, Nicolas V, Codogno P, Botti J (2013) Autophagy modulates cell migration and beta1 integrin membrane recycling. Cell Cycle 12:3317–3328. https://doi.org/10.4161/cc.26298
Acknowledgements
This study was supported by 2214/A-International Research Fellowship Program (for Doctorate Students) from The Scientific and Technological Research Council (TUBITAK).
Funding
This study was supported by 2214/A-International Research Fellowship Program (for Doctorate Students) from The Scientific and Technological Research Council (TUBITAK).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11010_2021_4063_MOESM1_ESM.tif
Fig. 1 Silencing UVRAG results in decrease colony formation. (a) and (b) ER+ cells MCF-7 and T47D were evaluated by colony formation as described in Materials and Methods. Student’s t test was conducted for statistical analysis. The results represented the mean ± standard deviation (SD) values obtained from at least three different experiments. Knockdown UVRAG does not affect apoptosis. (c) PARP apoptotic protein expression in MCF-7 cells was evaluated by western blot analysis after silencing UVRAG. β-actin was used as loading controls and all experiments were independently repeated three times (TIF 106 kb) (TIF 106 kb)
11010_2021_4063_MOESM2_ESM.tif
Fig. 2 The effect of UVRAG downregulation on cell motility activity. (a) and (b) MCF-7 and ZR-75.1 were seeded onto six-well plates. Then, Cells were exposed to UVRAG siRNAs or control siRNA (50 nM) for 72 h and motility/migration was evaluated by wound healing assay (TIF 263 kb) (TIF 264 kb)
Rights and permissions
About this article
Cite this article
Sencan, S., Tanriover, M., Ulasli, M. et al. UV radiation resistance-associated gene (UVRAG) promotes cell proliferation, migration, invasion by regulating cyclin-dependent kinases (CDK) and integrin-β/Src signaling in breast cancer cells. Mol Cell Biochem 476, 2075–2084 (2021). https://doi.org/10.1007/s11010-021-04063-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-021-04063-y