Abstract
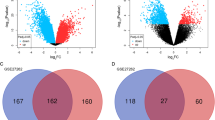
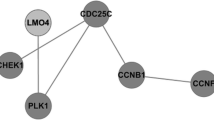
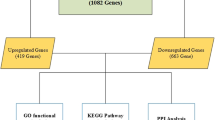
Lung adenocarcinoma (LUAD) accounts for the majority of cancer-related deaths worldwide. Our study identified key LUAD genes and their potential mechanism via bioinformatics analysis of public datasets. GSE10799, GSE40791, and GSE27262 microarray datasets were retrieved from the Gene Expression Omnibus (GEO) database. The RobustRankAggreg package was used to perform a meta-analysis, and 50 upregulated genes and 87 downregulated genes overlapped in three datasets. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID). Furthermore, protein–protein interaction (PPI) networks of the differentially expressed genes (DEGs) were built by the Search Tool for the Retrieval of Interacting Genes (STRING) and 22 core genes were identified by Molecular Complex Detection (MCODE) and visualized with Cytoscape. Subsequently, these core genes were analyzed by the Kaplan–Meier Plotter and Gene Expression Profiling Interactive Analysis (GEPIA). The results showed that all 22 genes were significantly associated with reduced survival rates. For GEPIA, the expression of only one gene was not significantly different between LUAD tissues and normal tissues. A KEGG pathway enrichment reanalysis of the 21 genes identified five key genes (CCNB1, BUB1B, CDC20, TTK, and MAD2L1) in the cell cycle pathway. Finally, the Comparative Toxicogenomics Database (CTD) website was used to explore the relationship between these key genes and certain drugs. Based on the bioinformatics analysis, five key genes were identified in LUAD, and drugs closely associated these genes can provide clues for the treatment and prognosis of LUAD.







Similar content being viewed by others
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61(2):69–90. https://doi.org/10.3322/caac.20107
Malvezzi M, Carioli G, Bertuccio P, Boffetta P, Levi F, La Vecchia C, Negri E (2017) European cancer mortality predictions for the year 2017, with focus on lung cancer. Ann Oncol 28(5):1117–1123. https://doi.org/10.1093/annonc/mdx033
Yao G, Chen K, Qin Y, Niu Y, Zhang X, Xu S, Zhang C, Feng M, Wang K (2019) Long non-coding RNA JHDM1D-AS1 interacts with DHX15 protein to enhance non-small-cell lung cancer growth and metastasis. Mol Ther Nucleic Acids 18:831–840. https://doi.org/10.1016/j.omtn.2019.09.028
Mao H (2019) Clinical relevance of mutant-allele tumor heterogeneity and lung adenocarcinoma. Ann Transl Med 7(18):432. https://doi.org/10.21037/atm.2019.08.112
Gan TQ, Chen WJ, Qin H, Huang SN, Yang LH, Fang YY, Pan LJ, Li ZY, Chen G (2017) Clinical value and prospective pathway signaling of microRNA-375 in lung adenocarcinoma: a study based on the Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO) and bioinformatics analysis. Med Sci Monit 23:2453–2464. https://doi.org/10.12659/msm.901460
Bedognetti D, Wang E, Sertoli MR, Marincola FM (2010) Gene-expression profiling in vaccine therapy and immunotherapy for cancer. Expert Rev Vaccines 9(6):555–565. https://doi.org/10.1586/erv.10.55
Coe BP, Chari R, Lockwood WW, Lam WL (2008) Evolving strategies for global gene expression analysis of cancer. J Cell Physiol 217(3):590–597. https://doi.org/10.1002/jcp.21554
Sotiriou C, Piccart MJ (2007) Taking gene-expression profiling to the clinic: when will molecular signatures become relevant to patient care? Nat Rev Cancer 7(7):545–553. https://doi.org/10.1038/nrc2173
Beane J, Spira A, Lenburg ME (2009) Clinical impact of high-throughput gene expression studies in lung cancer. J Thorac Oncol 4(1):109–118. https://doi.org/10.1097/JTO.0b013e31819151f8
Singhal S, Miller D, Ramalingam S, Sun SY (2008) Gene expression profiling of non-small cell lung cancer. Lung Cancer 60(3):313–324. https://doi.org/10.1016/j.lungcan.2008.03.007
Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Muertter RN, Holko M, Ayanbule O, Yefanov A, Soboleva A (2011) NCBI GEO: archive for functional genomics data sets – 10 years on. Nucleic Acids Res 39(Database issue):D1005–D1010. https://doi.org/10.1093/nar/gkq1184
Kolde R, Laur S, Adler P, Vilo J (2012) Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics 28(4):573–580. https://doi.org/10.1093/bioinformatics/btr709
Denny P, Feuermann M, Hill DP, Lovering RC, Plun-Favreau H, Roncaglia P (2018) Exploring autophagy with Gene Ontology. Autophagy 14(3):419–436. https://doi.org/10.1080/15548627.2017.1415189
Lindeberg M, Biehl BS, Glasner JD, Perna NT, Collmer A, Collmer CW (2009) Gene Ontology annotation highlights shared and divergent pathogenic strategies of type III effector proteins deployed by the plant pathogen Pseudomonas syringae pv tomato DC3000 and animal pathogenic Escherichia coli strains. BMC Microbiol 9(Suppl 1):S4. https://doi.org/10.1186/1471-2180-9-S1-S4
Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M (2019) New approach for understanding genome variations in KEGG. Nucleic Acids Res 47(D1):D590–D595. https://doi.org/10.1093/nar/gky962
Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K (2017) KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45(D1):D353–D361. https://doi.org/10.1093/nar/gkw1092
Cao D, Hustinx SR, Sui G, Bala P, Sato N, Martin S, Maitra A, Murphy KM, Cameron JL, Yeo CJ, Kern SE, Goggins M, Pandey A, Hruban RH (2004) Identification of novel highly expressed genes in pancreatic ductal adenocarcinomas through a bioinformatics analysis of expressed sequence tags. Cancer Biol Ther 3(11):1081–1089; discussion 1090–1081. https://doi.org/10.4161/cbt.3.11.1175
Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z (2017) GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 45(W1):W98–W102. https://doi.org/10.1093/nar/gkx247
Davis AP, King BL, Mockus S, Murphy CG, Saraceni-Richards C, Rosenstein M, Wiegers T, Mattingly CJ (2011) The comparative toxicogenomics database: update 2011. Nucleic Acids Res 39(Database issue):D1067–D1072. https://doi.org/10.1093/nar/gkq813
Fu X, Chen G, Cai ZD, Wang C, Liu ZZ, Lin ZY, Wu YD, Liang YX, Han ZD, Liu JC, Zhong WD (2016) Overexpression of BUB1B contributes to progression of prostate cancer and predicts poor outcome in patients with prostate cancer. Onco Targets Ther 9:2211–2220. https://doi.org/10.2147/OTT.S101994
Myslinski E, Gerard MA, Krol A, Carbon P (2007) Transcription of the human cell cycle regulated BUB1B gene requires hStaf/ZNF143. Nucleic Acids Res 35(10):3453–3464. https://doi.org/10.1093/nar/gkm239
Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, Kumar R, Jenkins RB, de Groen PC, Roche P, van Deursen JM (2004) BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet 36(7):744–749. https://doi.org/10.1038/ng1382
Wan X, Yeung C, Kim SY, Dolan JG, Ngo VN, Burkett S, Khan J, Staudt LM, Helman LJ (2012) Identification of FoxM1/Bub1b signaling pathway as a required component for growth and survival of rhabdomyosarcoma. Cancer Res 72(22):5889–5899. https://doi.org/10.1158/0008-5472.CAN-12-1991
Mansouri N, Movafagh A, Sayad A, Heidary Pour A, Taheri M, Soleimani S, Mirzaei HR, Alizadeh Shargh S, Azargashb E, Bazmi H, Allah Moradi H, Zandnia F, Hashemi M, Massoudi N, Mortazavi-Tabatabaei SA (2016) Targeting of BUB1b gene expression in sentinel lymph node biopsies of invasive breast cancer in Iranian female patients. Asian Pac J Cancer Prev 17(S3):317–321. https://doi.org/10.7314/apjcp.2016.17.s3.317
Hudler P, Britovsek NK, Grazio SF, Komel R (2016) Association between polymorphisms in segregation genes BUB1B and TTK and gastric cancer risk. Radiol Oncol 50(3):297–307. https://doi.org/10.1515/raon-2015-0047
Hahn MM, Vreede L, Bemelmans SA, van der Looij E, van Kessel AG, Schackert HK, Ligtenberg MJ, Hoogerbrugge N, Kuiper RP, de Voer RM (2016) Prevalence of germline mutations in the spindle assembly checkpoint gene BUB1B in individuals with early-onset colorectal cancer. Genes Chromosomes Cancer 55(11):855–863. https://doi.org/10.1002/gcc.22385
Liu AW, Cai J, Zhao XL, Xu AM, Fu HQ, Nian H, Zhang SH (2009) The clinicopathological significance of BUBR1 overexpression in hepatocellular carcinoma. J Clin Pathol 62(11):1003–1008. https://doi.org/10.1136/jcp.2009.066944
Mussnich P, Raverot G, Jaffrain-Rea ML, Fraggetta F, Wierinckx A, Trouillas J, Fusco A, D’Angelo D (2015) Downregulation of miR-410 targeting the cyclin B1 gene plays a role in pituitary gonadotroph tumors. Cell Cycle 14(16):2590–2597. https://doi.org/10.1080/15384101.2015.1064207
Hwang A, McKenna WG, Muschel RJ (1998) Cell cycle-dependent usage of transcriptional start sites. A novel mechanism for regulation of cyclin B1. J Biol Chem 273(47):31505–31509. https://doi.org/10.1074/jbc.273.47.31505
Pandey JP, Kistner-Griffin E, Namboodiri AM, Iwasaki M, Kasuga Y, Hamada GS, Tsugane S (2014) Higher levels of antibodies to the tumour-associated antigen cyclin B1 in cancer-free individuals than in patients with breast cancer. Clin Exp Immunol 178(1):75–78. https://doi.org/10.1111/cei.12385
Yuan J, Yan R, Kramer A, Eckerdt F, Roller M, Kaufmann M, Strebhardt K (2004) Cyclin B1 depletion inhibits proliferation and induces apoptosis in human tumor cells. Oncogene 23(34):5843–5852. https://doi.org/10.1038/sj.onc.1207757
Suzuki T, Urano T, Miki Y, Moriya T, Akahira J, Ishida T, Horie K, Inoue S, Sasano H (2007) Nuclear cyclin B1 in human breast carcinoma as a potent prognostic factor. Cancer Sci 98(5):644–651. https://doi.org/10.1111/j.1349-7006.2007.00444.x
Zhou L, Li J, Zhao YP, Cui QC, Zhou WX, Guo JC, You L, Wu WM, Zhang TP (2014) The prognostic value of Cyclin B1 in pancreatic cancer. Med Oncol 31(9):107. https://doi.org/10.1007/s12032-014-0107-4
Takeno S, Noguchi T, Kikuchi R, Uchida Y, Yokoyama S, Muller W (2002) Prognostic value of cyclin B1 in patients with esophageal squamous cell carcinoma. Cancer 94(11):2874–2881. https://doi.org/10.1002/cncr.10542
Chu Z, Zhang X, Li Q, Hu G, Lian CG, Geng S (2019) CDC20 contributes to the development of human cutaneous squamous cell carcinoma through the Wnt/betacatenin signaling pathway. Int J Oncol 54(5):1534–1544. https://doi.org/10.3892/ijo.2019.4727
Bharadwaj R, Yu H (2004) The spindle checkpoint, aneuploidy, and cancer. Oncogene 23(11):2016–2027. https://doi.org/10.1038/sj.onc.1207374
Shang G, Ma X, Lv G (2018) Cell division cycle 20 promotes cell proliferation and invasion and inhibits apoptosis in osteosarcoma cells. Cell Cycle 17(1):43–52. https://doi.org/10.1080/15384101.2017.1387700
Li Y, Benezra R (1996) Identification of a human mitotic checkpoint gene: hsMAD2. Science 274(5285):246–248. https://doi.org/10.1126/science.274.5285.246
Abal M, Obrador-Hevia A, Janssen KP, Casadome L, Menendez M, Carpentier S, Barillot E, Wagner M, Ansorge W, Moeslein G, Fsihi H, Bezrookove V, Reventos J, Louvard D, Capella G, Robine S (2007) APC inactivation associates with abnormal mitosis completion and concomitant BUB1B/MAD2L1 up-regulation. Gastroenterology 132(7):2448–2458. https://doi.org/10.1053/j.gastro.2007.03.027
Vleugel M, Hoek TA, Tromer E, Sliedrecht T, Groenewold V, Omerzu M, Kops GJ (2015) Dissecting the roles of human BUB1 in the spindle assembly checkpoint. J Cell Sci 128(16):2975–2982. https://doi.org/10.1242/jcs.169821
Ko YH, Roh JH, Son YI, Chung MK, Jang JY, Byun H, Baek CH, Jeong HS (2010) Expression of mitotic checkpoint proteins BUB1B and MAD2L1 in salivary duct carcinomas. J Oral Pathol Med 39(4):349–355. https://doi.org/10.1111/j.1600-0714.2009.00835.x
Wang Z, Katsaros D, Shen Y, Fu Y, Canuto EM, Benedetto C, Lu L, Chu WM, Risch HA, Yu H (2015) Biological and clinical significance of MAD2L1 and BUB1, genes frequently appearing in expression signatures for breast cancer prognosis. PLoS One 10(8):e0136246. https://doi.org/10.1371/journal.pone.0136246
Xie Y, Wang A, Lin J, Wu L, Zhang H, Yang X, Wan X, Miao R, Sang X, Zhao H (2017) Mps1/TTK: a novel target and biomarker for cancer. J Drug Target 25(2):112–118. https://doi.org/10.1080/1061186X.2016.1258568
Jemaa M, Galluzzi L, Kepp O, Senovilla L, Brands M, Boemer U, Koppitz M, Lienau P, Prechtl S, Schulze V, Siemeister G, Wengner AM, Mumberg D, Ziegelbauer K, Abrieu A, Castedo M, Vitale I, Kroemer G (2013) Characterization of novel MPS1 inhibitors with preclinical anticancer activity. Cell Death Differ 20(11):1532–1545. https://doi.org/10.1038/cdd.2013.105
Maia AR, de Man J, Boon U, Janssen A, Song JY, Omerzu M, Sterrenburg JG, Prinsen MB, Willemsen-Seegers N, de Roos JA, van Doornmalen AM, Uitdehaag JC, Kops GJ, Jonkers J, Buijsman RC, Zaman GJ, Medema RH (2015) Inhibition of the spindle assembly checkpoint kinase TTK enhances the efficacy of docetaxel in a triple-negative breast cancer model. Ann Oncol 26(10):2180–2192. https://doi.org/10.1093/annonc/mdv293
Liang XD, Dai YC, Li ZY, Gan MF, Zhang SR, Yin P, Lu HS, Cao XQ, Zheng BJ, Bao LF, Wang DD, Zhang LM, Ma SL (2014) Expression and function analysis of mitotic checkpoint genes identifies TTK as a potential therapeutic target for human hepatocellular carcinoma. PLoS One 9(6):e97739. https://doi.org/10.1371/journal.pone.0097739
Liu X, Liao W, Yuan Q, Ou Y, Huang J (2015) TTK activates Akt and promotes proliferation and migration of hepatocellular carcinoma cells. Oncotarget 6(33):34309–34320. https://doi.org/10.18632/oncotarget.5295
Tannous BA, Kerami M, Van der Stoop PM, Kwiatkowski N, Wang J, Zhou W, Kessler AF, Lewandrowski G, Hiddingh L, Sol N, Lagerweij T, Wedekind L, Niers JM, Barazas M, Nilsson RJ, Geerts D, De Witt Hamer PC, Hagemann C, Vandertop WP, Van Tellingen O, Noske DP, Gray NS, Wurdinger T (2013) Effects of the selective MPS1 inhibitor MPS1-IN-3 on glioblastoma sensitivity to antimitotic drugs. J Natl Cancer Inst 105(17):1322–1331. https://doi.org/10.1093/jnci/djt168
Elbaz HA, Stueckle TA, Wang HY, O'Doherty GA, Lowry DT, Sargent LM, Wang L, Dinu CZ, Rojanasakul Y (2012) Digitoxin and a synthetic monosaccharide analog inhibit cell viability in lung cancer cells. Toxicol Appl Pharmacol 258(1):51–60. https://doi.org/10.1016/j.taap.2011.10.007
Yan KH, Yao CJ, Chang HY, Lai GM, Cheng AL, Chuang SE (2010) The synergistic anticancer effect of troglitazone combined with aspirin causes cell cycle arrest and apoptosis in human lung cancer cells. Mol Carcinog 49(3):235–246. https://doi.org/10.1002/mc.20593
Funding
The research was supported by grants from the National Natural Science Foundation of China (No. 81673153 and No. 81973034).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest with the publication of the manuscript.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, ., Zhou, Z., Chen, L. et al. Identification of key genes and biological pathways in lung adenocarcinoma via bioinformatics analysis. Mol Cell Biochem 476, 931–939 (2021). https://doi.org/10.1007/s11010-020-03959-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-020-03959-5