Abstract
Several reports demonstrated the direct contribution of cytochrome P450 1B1 (CYP1B1) enzyme and its associated cardiotoxic mid-chain, hydroxyeicosatetraenoic acid (HETEs) metabolites in the development of cardiac hypertrophy. Resveratrol is commercially available polyphenol that exerts beneficial effects in wide array of cardiovascular diseases including cardiac hypertrophy, myocardial infarction and heart failure. Nevertheless, the underlying mechanisms responsible for these effects are not fully elucidated. Since resveratrol is a well-known CYP1B1 inhibitor, the purpose of this study is to test whether resveratrol attenuates angiotensin II (Ang II)-induced cellular hypertrophy through inhibition of CYP1B1/mid-chain HETEs mechanism. RL-14 and H9c2 cells were treated with vehicle or 10 μM Ang II in the absence and presence of 2, 10 or 50 μM resveratrol for 24 h. Thereafter, the level of mid-chain HETEs was determined using liquid chromatography–mass spectrometry (LC/MS). Hypertrophic markers and CYP1B1 gene expression and protein levels were measured using real-time PCR and Western blot analysis, respectively. Our results demonstrated that resveratrol, at concentrations of 10 and 50 μM, was able to attenuate Ang-II-induced cellular hypertrophy as evidenced by substantial inhibition of hypertrophic markers, β-myosin heavy chain (MHC)/α-MHC and atrial natriuretic peptide. Ang II significantly induced the protein expression of CYP1B1 and increased the metabolite formation rate of its associated mid-chain HETEs. Interestingly, the protective effect of resveratrol was associated with a significant decrease of CYP1B1 protein expression and mid-chain HETEs. Our results provided the first evidence that resveratrol protects against Ang II-induced cellular hypertrophy, at least in part, through CYP1B1/mid-chain HETEs-dependent mechanism.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pathological cardiac hypertrophy is a complex condition that is characterized by increased thickness of the cardiac muscle and a reduction of the size of the chambers of the heart, representing a major risk factor for developing most phenotypes of heart failure [1, 2]. Heart failure (HF) remains a global pandemic that represents a leading cause of morbidity and mortality worldwide. In spite of considerable gains in the treatment options, there are yet cases where HF patients continue to be symptomatic, posing a major burden on the health care system [3, 4]. According to Heart and Stroke Foundation of Canada, HF is affecting more than 600,000 Canadians with 50,000 new cases being diagnosed with the disease annually and it is responsible for an estimated health expenditure of around $2.8 billion per year as a direct cost [5]. Searching for possible treatments that halt the development and progression of cardiac hypertrophy is regarded as an important step in the prevention of a major risk factor of heart failure and will allow for designing novel therapeutic modalities.
A plethora of molecular mechanisms are involved in the pathogenesis of cardiac hypertrophy and the consequent cardiac remodeling. One of these mechanisms is cytochrome P450 (CYP) enzymes and their associated arachidonic acid (AA) metabolites [6, 7]. In the heart, CYP enzymes metabolize AA into either cardioprotective metabolites, such as epoxyeicosatrienoic acids (EETs) and 19-hydroxyeicosatetraenoic acid (19-HETE), or cardiotoxic metabolites exemplified as mid-chain HETEs and 20-HETE [8, 9]. Recently, cardiac CYP1B1 and its associated mid-chain HETEs, typified by 5-, 8-, 9-, 11-, 12- and 15-HETE, have received a particular interest as a potential druggable target. Mid-chain HETEs are group of biologically active eicosanoids that are produced from the metabolism of AA by both lipoxygenase- and CYP-catalyzed bis-allylic oxidation reaction [10, 11].
A growing body of evidence demonstrated that mid-chain HETEs possess cardiotoxic effects and they are involved in the pathogenesis of cardiovascular diseases (CVD) including atherosclerosis, hypertension, cardiac hypertrophy and HF [12,13,14]. Several reports showed that overexpression of CYP1B1 in the cardiac muscle caused an increase in the level of cardiotoxic mid-chain HETEs leading to deterioration of the cardiac histo-architecture and function [12, 15, 16]. On the contrary, strategies to mitigate CYP1B1 activity either through pharmacological inhibition or genetic deletion have been employed to protect against CVD. 2,3′,4,5′-tetramethoxystilbene (TMS), a well-known CYP1B1 inhibitor, was able to significantly decrease the level of mid-chain HETEs and protect against angiotensin II (Ang II)-induced cardiac hypertrophy [17]. In addition, CYP1B1 genetic disruption resulted in a reduction of cardiac hypertrophy in Ang II-induced model of hypertension in mice [10].
Resveratrol is a naturally occurring polyphenol that is found in many food sources and beverages including black grapes, peanuts, blueberries and red wine and as a commercially available dietary supplement [18]. Resveratrol possesses anti-oxidant, anti-inflammatory, anti-apoptotic and anti-fibrotic effects. Of interest, resveratrol has been shown to improve the myocardial energetics and is experimentally used for prevention and treatment of multiple CVD [19, 20]. Resveratrol has been shown to exert an inhibitory effect on recombinant CYP1B1 supersomes with IC50 = 11.2 µM [21]. In addition, it inhibited TCDD-mediated induction of CYP1B1 in cultured human mammary epithelial cells and human breast cancer cell line MCF-7 [22, 23]. Several reports have demonstrated that resveratrol protects against cardiac hypertrophy through modulation of various pathways [24,25,26]. However, none of these studies investigated the effects of resveratrol on CYP1B1. Therefore, the purpose of this study is to investigate whether resveratrol protects against Ang II-induced cellular hypertrophy through inhibition of CYP1B1/mid-chain HETEs mechanism.
Materials and methods
Materials
Resveratrol was purchased from Sigma-Aldrich Co. (St. Louis, MO, USA). 15-HETE-D8 was purchased from Cayman Chemical (Ann Arbor, MI, USA). Dulbecco’s Modified Eagle’s Medium/F-12 (DMEM/F-12) and DMEM were obtained from Gibco, Life Technologies (Grand Island, NY, USA). Fetal bovine serum was purchased from Sigma-Aldrich Co. (St. Louis, MO, USA). TRIzol reagent was purchased from Invitrogen Co. (Carlsbad, CA, USA). High-Capacity cDNA Reverse Transcription kit and SYBR® Green PCR Master Mix were purchased from Applied Biosystems (Foster City, CA, USA). Real-time PCR primers were prepared by Integrated DNA Technologies (Coralville, IA, USA). Immun-Blot® PVDF membrane was purchased from Bio-Rad Laboratories (Hercules, CA, USA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mouse monoclonal was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). Recombinant monoclonal anti-CYP1B1 antibody [ab185954] was purchased from abcam (Toronto, ON). Chemiluminescence Western blotting detection reagents (ECL) were obtained from GE Healthcare Life Sciences (Pittsburgh, PA, USA). All of the other chemicals used were obtained from Fisher Scientific Co. (Toronto, ON).
Cell culture
Human cardiomyocyte (RL-14) cells and rat cardiomyoblast (H9c2) cell lines were obtained from American Type Culture Collection (ATCC) (Manassas, VA, USA). RL-14 cells were grown in DMEM/F-12, with phenol red supplemented with 12.5% fetal bovine serum, 20 μM l-glutamine, 1% penicillin–streptomycin. H9c2 cells were grown in DMEM containing 10% heat-inactivated fetal bovine serum, and 1% penicillin–streptomycin. Cells were grown in 75 cm2 cell culture flasks at 37 °C in a 5% CO2 humidified environment.
Chemical treatments
Cells were treated with vehicle (serum-free DMEM/F-12 containing 0.1% DMSO for RL-14 cells and serum-free DMEM containing 0. 05% dimethyl sulfoxide (DMSO) for H9c2 cells), 10 μM Ang II, 10 μM Ang II in combination with (2, 10 or 50 μM resveratrol) or (2, 10 or 50 μM resveratrol alone) in serum-free medium (SFM) for the time indicated for each experiment as described in the figure legends. Resveratrol was prepared as a stock solution in DMSO and maintained in DMSO at − 20 °C until use. The treatment of cells was performed in the corresponding culture media in 12-well cell culture plates for RNA assay and 6-well cell culture plates for protein and determination of AA metabolites assays. Ang II stock solution was prepared in saline and maintained at − 20 °C until use. Diluted solutions of Ang II and resveratrol were freshly prepared in SFM before each experiment. In all experiments, the DMSO concentration did not exceed 0.05% (v/v).
Cell viability analysis
MTT assay was used to determine the viability of RL-14 and H9c2 cells. The main principle of this assay is the ability of viable cells to reduce tetrazolium dye MTT 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide to its insoluble colored formazan crystals. Briefly, cells were seeded in 96-well plates, treated with varying concentrations of resveratrol, incubated for 24 h at 37 °C in a 5% CO2 humidified incubator, incubated with 20 μl/well MTT (1.2 mM) dissolved in phosphate-buffered saline (PBS; pH 7.4) for 3 h at 37 °C, and then 150 μl/well isopropyl alcohol to dissolve the formazan for 10 min at room temperature. The optical density was assessed at a wavelength of 550 nm using the Bio-Tek Synergy H1 Hybrid Multi-Mode Microplate Reader (Bio-Tek Instruments, Winooski, VT, USA). The cell viability was presented as a percentage of the control mean absorbance value.
RNA extraction and cDNA synthesis
RL-14 and H9c2 cells were seeded in 12-well plates and treated with the test compounds for 24 h. Thereafter, cells were collected and total RNA was isolated using TRIzol reagent (Invitrogen®) according to the manufacturer’s instructions, and quantified by measuring the absorbance at 260 nm while the purity was determined by measuring the 260/280 ratio (> 1.8). Afterward, first-strand cDNA synthesis was carried out using the High-Capacity cDNA reverse transcription kit (Applied Biosystems) according to the manufacturer’s instructions. In brief, 1.5 μg of total RNA from each sample was added to a mix of 2.0 μl 10 × RT buffer, 0.8 μl 25 × dNTP mix (100 mM), 2.0 μl 10 × reverse transcriptase random primers, 1.0 μl MultiScribe™ reverse transcriptase enzyme and 4.2 μl nuclease-free water. The final reaction mixture was kept at 25 °C for 10 min, heated to 37 °C for 120 min, heated for 85 °C for 5 min and finally cooled to 4 °C.
Real-time polymerase chain reaction (real-time PCR) for quantification of mRNA expression
Real-time PCR was utilized to quantitatively determine specific mRNA expression of different targets by subjecting the resultant cDNA to PCR amplification using 96-well optical reaction plates in the ABI Prism 7500 System (Applied Biosystems). The reaction mixture (25 μl) contained 0.1 μl of 10 μM forward primer and 0.1 μl of 10 μM reverse primers (40 nM final concentration of each primer), 12.5 μl of SYBR Green Universal Master mix, 11.05 μl of RNAse/DNAse-free water, and 1.25 μl of cDNA sample. Human primer sequences for CYP1B1, atrial natriuretic peptide (ANP), α-myosin heavy chain (α-MHC), β-myosin heavy chain (β-MHC), and β-actin as well as rat primer sequences for CYP1B1, ANP, α-MHC, β-MHC, and β-actin are listed in Table 1. Primers were purchased from Integrated DNA technologies IDT (Coralville, IA, USA). Analysis of the real-time PCR data was carried out using the relative gene expression (i.e., ΔΔCT) method. In brief, the fold change in the level of target genes between treated and untreated cells, corrected for the level of housekeeping gene (β-actin), was determined using the following equation: fold change = 2−Δ(ΔCt), where ΔCt = Ct(target) − Ct(β-actin) and Δ(ΔCt) = ΔCt(treated) − ΔCt(untreated). The thermal cycle parameters were as follow: initiation of the reaction at 95 °C for 10 min and 40 cycles of denaturation (95 °C, 15 s) and combined annealing/extension (60 °C, 60 s).
Protein extraction
RL-14 and H9c2 cells were grown in 6-well plates and incubated with the test compounds for 24 h. Thereafter, lysis buffer containing 50 mM HEPES, 0.5 M sodium chloride, 1.5 mM magnesium chloride, 1 mM EDTA, 10% (v/v) glycerol, 1% Triton X-100, and 5 μl/ml protease inhibitor cocktail was added to collect the cells. Cell lysates were incubated on ice for 1 h to prepare the cell homogenates while they are sporadically vortexed every 10 min, followed by centrifugation at 12,000×g for 10 min at 4 °C. Supernatant of total cellular lysate was collected and maintained at − 80 °C. Subsequently, Lowry assay was carried out to determine the concentration of protein using bovine serum albumin as a standard [27].
Western blot analysis
Western blot analysis was performed according to previously detailed assay [16]. Briefly, total cell lysates (50 μg) were separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE), samples were undergone electrophoresis at 120 V for 2 h and separated proteins were transferred onto Immun-Blot® PVDF membrane. Afterward, protein membranes were blocked overnight at 4 °C using blocking solution containing 0.15 M sodium chloride, 3 mM potassium chloride, 25 mM Tris-base, 5% skim milk, 2% bovine serum albumin, and 0.5% Tween-20. Following blocking, the blots were subjected to washing cycles 3 times for 30 min with Tris-buffered saline (TBS)–Tween-20. The blots were subsequently incubated for 2 h at 4 °C with primary antibodies in TBS solution (0.05% (v/v) Tween-20, 0.02% sodium azide). Incubation with secondary antibodies (peroxidase-conjugated IgG) in blocking solution was performed for 45 min at room temperature. Visualization of the bands was carried out using the enhanced chemiluminescence method according to the manufacturer’s guide (GE Healthcare Life Sciences). ImageJ software (National Institutes of Health, Bethesda, MD, USA; https://rsb.info.nih.gov/ij) was used to quantify the intensity of the protein bands in relation to the signals acquired from GAPDH loading control. Data, given in the figures, are represented as relative protein intensity (%) + SEM, as compared to control group.
Metabolism of AA by H9c2 and RL-14 cells
To investigate the effect of resveratrol on mid-chain HETE metabolites, RL-14 and H9c2 cells were treated, as previously described, for 24 h and then the cells were incubated with 50 μM AA for 3 h. Extraction of AA metabolites was carried out using ethyl acetate and dried using speed vacuum (Savant, Farmingdale, NY, USA). The resultant metabolites were analyzed using liquid chromatography–electrospray ionization mass spectrometry (LC–ESI–MS) (Waters Micromass ZQ 4000 spectrometer) method.
Apparatus and chromatographic conditions
The analysis of mid-chain HETE metabolites was performed using LC–ESI–MS as previously described [28]. Briefly, negative ionization mode was the mode of the mass spectrometer with single ion monitoring: m/z = 319 for mid-chain HETE metabolites, and m/z = 327 for internal standard, 15-HETE-D8. The nebulizer gas was supplied from an in house nitrogen source with high purity. The source temperature was set to of 150 °C, and voltage of the capillary and cone were 3.51 kV and 25 V, respectively. A gradient separation was performed on a reverse phase C18 column (Alltima HP, 150 × 2.1 mm) at 35 °C. The mobile phase (A) was composed of water with 0.01% formic acid and 0.005% triethylamine (v/v), whereas mobile phase (B) consisted of 8% methanol, 8% isopropanol, and 84% acetonitrile with 0.01% formic acid and 0.005% triethylamine (v/v). Samples were subjected to linear gradient elution at a flow rate of 200 μl/min, as follows: 60 to 48% in 4 min, held isocratically at 48% for 24 min, 48 to 35% in 11 min, 35 to 0% in 11 min, and finally held isocratically at 0% for 7 min of mobile phase A.
Statistical analysis
All results are presented as the mean ± SEM. Multiple group comparisons was performed using one-way analysis of variance (ANOVA) followed by the Student–Newman–Keuls as a post hoc test. Differences between means were considered significant at p < 0.05. All analyses were performed using SigmaPlot® for Windows (Systat Software, San Jose, CA, USA).
Results
Effect of resveratrol on cell viability in RL-14 and H9c2 cells
MTT assay was used to assess the effect of different concentrations of resveratrol on cell viability. RL-14 and H9c2 cells were grown to 80–90% confluency in 96-well culture plates and treated for 24 h with increasing concentrations of resveratrol (0, 1, 10, 25, 50 or 100 µM). Cells in the control group were treated with SFM without resveratrol. After incubation of cells with MTT for 3 h, MTT is reduced by viable cells to produce formazan dye which is then solubilized using isopropyl alcohol. Data in Fig. 1a, b showed that resveratrol, at all concentrations used, did not significantly alter the cell viability of both cell lines (measured cell viability was more than 90%). Therefore, 2, 10 and 50 µM resveratrol concentrations were selected to perform all consequent experiments in the current study based on MTT assay results.
Effect of resveratrol on cell viability in RL-14 and H9c2 cells. RL-14 (a) and H9c2 (b) cells were treated for 24 h with 1–100 μM of resveratrol. Cell cytotoxicity was assessed using the MTT assay. Data are presented as the percentage of control (set at 100%) ± SEM (n = 4). Data were analyzed using one-way ANOVA followed by Student–Newman–Keuls as post hoc test
Effect of resveratrol on Ang II-induced cellular hypertrophy in RL-14 and H9c2 cells
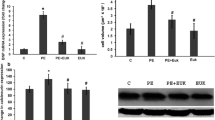
To examine the potential protective effect of resveratrol against the development of cellular hypertrophy, RL-14 and H9c2 cells were treated with vehicle, 10 μM Ang II, 10 μM Ang II in combination with resveratrol (2, 10 or 50 μM) or resveratrol alone (2, 10 or 50 μM) in SFM. The results showed that incubation of cells with 10 μM Ang II for 24 h resulted in cellular hypertrophy as demonstrated by significant increase in β/α-MHC ratio and ANP by approximately 123% and 78%, respectively in RL-14 cells, compared to control group (Fig. 2a). Also, in H9c2 cells, Ang II caused significant elevation of β/α-MHC ratio and ANP by approximately 226% and 270%, respectively, compared to control group (Fig. 2b). While low concentration of resveratrol (2 μM) was not able to revert Ang II-induced effects, co-treatment with higher concentration of 10 and 50 μM was able to significantly protect against Ang II-mediated elevation of β/α-MHC ratio and ANP to approximately control levels in both cell lines. Notably, treatment of cells with low and high concentrations of resveratrol alone did not have any significant effects on hypertrophic markers. These results indicate that resveratrol protects against Ang II-induced cellular hypertrophy as evidenced by a reduction in hypertrophic markers and prompted us to further investigate the effects of resveratrol on CYP1B1 and its cardiotoxic mid-chain HETE metabolites.
Effect of resveratrol on Ang II-mediated induction of hypertrophic markers in RL-14 and H9c2 cells. RL-14 (a) and H9c2 (b) cells were treated for 24 h with vehicle, 10 μM Ang II, 10 μM Ang II in combination with (2, 10 or 50 μM resveratrol) or (2, 10 or 50 μM resveratrol alone) in SFM. Total RNA was isolated using TRIzol, the mRNA expression levels of β-MHC/α-MHC and ANP were assessed using real-time PCR and their levels were normalized to β-actin housekeeping gene. The results are presented as the mean and SEM based on at least 3 individual experiments. Data were analyzed using one-way ANOVA followed by Student–Newman–Keuls as post hoc test. +p < 0.05 significantly different from control group. *p < 0.05 significantly different from Ang II-treated group
Effect of resveratrol on the mRNA and protein expression of CYP1B1 in RL-14 and H9c2 cells
We investigated the effect of low and high concentrations of resveratrol on mRNA and protein expression levels of CYP1B1 in RL-14 and H9c2 cells and the results showed that while Ang II (10 μM) had no significant effect on CYP1B1 mRNA level in comparison to control group in both cell lines, co-treatment with Ang II and resveratrol (2, 10 or 50 μM) or resveratrol alone (2, 10 or 50 μM) significantly decreased the transcription level of CYP1B1, compared to Ang II group, in both cell lines (Figs. 3a, 4a). In RL-14 cells, resveratrol (2, 10 or 50 μM) in combination with Ang II decreased CYP1B1 mRNA expression by approximately 40%, 81% or 89%, respectively. While 2, 10 or 50 μM resveratrol alone decreased the expression of CYP1B1 by approximately 60%, 80% or 86%, respectively. In H9c2 cells, resveratrol (2, 10 or 50 μM) in combination with Ang II decreased CYP1B1 mRNA expression by approximately 35%, 74% or 88%, respectively. However, 2, 10 or 50 μM resveratrol alone decreased the expression by approximately 64%, 88% or 89%, respectively.
Effect of resveratrol on mRNA expression and protein expression levels of CYP1B1 in RL-14 cells. RL-14 cells were treated for 24 h with vehicle, 10 μM Ang II, 10 μM Ang II in combination with (2, 10 or 50 μM resveratrol) or (2, 10 or 50 μM resveratrol alone) in SFM. CYP1B1 mRNA expression (a) and protein expression levels (b) were determined using real-time PCR and Western blot analysis, respectively. For real-time PCR, total RNA was isolated using TRIzol reagent, the mRNA level was quantified and its level was normalized to β-actin housekeeping gene. For Western blot analysis, protein levels were detected using the enhanced chemiluminescence method. The intensity of protein band was normalized to the signals obtained for β-actin or GAPDH protein and quantified using ImageJ®. The results are presented as the mean and SEM based on at least 3 individual experiments. Data were analyzed using one-way ANOVA followed by Student–Newman–Keuls as post hoc test. +p < 0.05 significantly different from control group. *p < 0.05 significantly different from Ang II-treated group
Effect of resveratrol on mRNA expression and protein expression levels of CYP1B1 in H9c2 cells. H9c2 cells were treated for 24 h with vehicle, 10 μM Ang II, 10 μM Ang II in combination with (2, 10 or 50 μM resveratrol) or (2, 10 or 50 μM resveratrol alone) in SFM. CYP1B1 mRNA expression (a) and protein expression levels (b) were determined using real-time PCR and Western blot analysis, respectively. For real-time PCR, total RNA was isolated using TRIzol reagent, the mRNA level was quantified and its level was normalized to β-actin housekeeping gene. For Western blot analysis, protein levels were detected using the enhanced chemiluminescence method. The intensity of protein band was normalized to the signals obtained for β-actin or GAPDH protein and quantified using ImageJ®. The results are presented as the mean and SEM based on at least 3 individual experiments. Data were analyzed using one-way ANOVA followed by Student–Newman–Keuls as post hoc test. +p < 0.05 significantly different from control group. *p < 0.05 significantly different from Ang II-treated group
We examined whether different concentrations of resveratrol have a protective effect against Ang II-induced cellular hypertrophy through inhibiting the protein expression of CYP1B1. For this reason, Western blot analysis was performed and the results showed that the protein expression of CYP1B1 was significantly increased by Ang II treatment in comparison to control group in RL-14 and H9c2 cells, by approximately 570% and 430%, respectively (Figs. 3b, 4b). In RL-14 cells, low and high concentrations of resveratrol (2, 10 or 50 μM) in combination with Ang II significantly decreased CYP1B1 protein expression by approximately 19%, 28% or 58%, respectively, compared to Ang II group. Interestingly, 2, 10 or 50 μM resveratrol alone induced CYP1B1 protein expression by approximately 2.6-, 2.6- or 2-folds, respectively. In H9c2 cells, low concentration of resveratrol (2 μM) did not significantly alter CYP1B1 protein level compared to Ang II group. However, high concentrations of resveratrol (10 or 50 μM) in combination with Ang II significantly decreased CYP1B1 protein expression by approximately 31% or 33%, respectively, compared to Ang II group. Intriguingly, 2, 10 or 50 μM resveratrol alone increased CYP1B1 protein expression by approximately 1.7-, 1.6- or 2-folds, respectively.
Effect of low and high concentrations of resveratrol on the formation of mid-chain HETEs in RL-14 and H9c2 cells
To investigate the effect of low and high concentrations of resveratrol on the formation of mid-chain HETEs metabolites, RL-14 and H9c2 cells were treated for 24 h with vehicle, 10 μM Ang II or 10 μM Ang II in combination with (2, 10 or 50 μM resveratrol). Thereafter, the cells were incubated with 50 μM AA for 3 h and mid-chain HETE metabolites were analyzed using LC–ESI–MS. The results showed that treatment of RL-14 cells with Ang II significantly increased the metabolite formation rate of 5-, 8-, 9-, 12- and 15-HETE by approximately 427%, 303%, 279%, 277% and 425%, respectively, while it had no significant effect on 11-HETE (Fig. 5a, b). Resveratrol at concentration of 2 μM was able to decrease Ang II-mediated induction of 5- and 12-HETE by approximately 51% and 53%, respectively, compared to Ang II-treated group. While it was not able to reverse this increase in case of 8-, 9-, and 15-HETE (Fig. 5a, b). On the other hand, higher doses of resveratrol significantly protected against the increase in cardiotoxic mid-chain HETE metabolites. Resveratrol at concentration of 10 μM significantly inhibited Ang II-mediated increase of the metabolite formation rate of 5-, 8-, 9-, 12- and 15-HETE by approximately 78%, 73%, 65%, 64% and 62%, respectively, compared to Ang II-treated group (Fig. 5a, b). Also, the highest concentration of resveratrol (50 μM) was able to decrease Ang II-mediated increase of the metabolite formation rate of 5-, 8-, 9-, 12- and 15-HETE by approximately 66%, 73%, 68%, 67% and 66%, respectively, compared to Ang II-treated group (Fig. 5a, b).
Effect of resveratrol on mid-chain HETE metabolite formation rate in RL-14 cells. RL-14 cells were treated for 24 h with vehicle, 10 μM Ang II and 10 μM Ang II in combination with (2, 10 or 50 μM resveratrol). 5-, 8-, 9-HETE (a) and 11-, 12-, 15-HETE (b) metabolites were measured using LC–ESI–MS. The results are presented as the mean and SEM (n = 3). Data were analyzed using one-way ANOVA followed by Student–Newman–Keuls as post hoc test. +p < 0.05 significantly different from control group. *p < 0.05 significantly different from Ang II-treated group
To further confirm the results in another species, we have performed the metabolic analysis study in rat H9c2 cells. The results showed that treatment of H9c2 cells with 10 μM Ang II significantly increased the metabolite formation rate of 5-, 8-, 12- and 15-HETE by approximately 362%, 301%, 303% and 344%, respectively, while it had no significant effect on 9- or 11-HETE (Fig. 6a, b). Resveratrol at concentration of 2 μM was only able to decrease Ang II-mediated increase of 15-HETE by approximately 34%, compared to Ang II-treated group. While it was not able to reverse, this increase in case of 5-, 8- and 12-HETE (Fig. 6a, b). Interestingly, higher concentrations of resveratrol also significantly inhibited Ang II-mediated increase in cardiotoxic mid-chain HETE metabolites. Resveratrol at concentration of 10 μM significantly inhibited Ang II-mediated increase of the metabolite formation rate of 5-, 8-, 12- and 15-HETE by approximately 65%, 62%, 65% and 64%, respectively, compared to Ang II-treated group (Fig. 6a, b). Moreover, the highest concentration of resveratrol (50 μM) was able to decrease Ang II-mediated increase of the metabolite formation rate of 5-, 8-, 12- and 15-HETE by approximately 67%, 69%, 68% and 68%, respectively, compared to Ang II-treated group (Fig. 6a, b).
Effect of resveratrol on mid-chain HETE metabolite formation rate in H9c2 cells. H9c2 cells were treated for 24 h with vehicle, 10 μM Ang II and 10 μM Ang II in combination with (2, 10 or 50 μM resveratrol). 5-, 8-, 9-HETE (a) and 11-, 12-, 15-HETE (b) metabolites were measured using LC–ESI–MS. The results are presented as the mean and SEM (n = 3). Data were analyzed using one-way ANOVA followed by Student–Newman–Keuls as post hoc test. +p < 0.05 significantly different from control group. *p < 0.05 significantly different from Ang II-treated group
Discussion
According to World Health Organization (WHO), CVD are the leading cause of death worldwide, they are responsible for the death of approximately 17.9 million people each year representing 31% of all deaths globally [28]. Accumulating studies have demonstrated that CYP enzymes and their associated AA metabolites play a central role in the maintenance of cardiovascular health and also in the pathogenesis of CVD [29, 30]. Interestingly, CYP1B1 and its associated cardiotoxic mid-chain HETE metabolites have been given a considerable attention due to their evident role in the pathogenesis of CVD such as pathological cardiac hypertrophy, atherosclerosis, hypertension and HF [31, 32]. Moreover, several studies have established an inverse relationship between developing CVD and the dietary consumption of vegetables and fruits. These protective effects have been attributed to polyphenols including flavonoids (such as resveratrol) and stilbenoids that are naturally present in high concentration in heart-healthy diets. Nevertheless, the molecular cardioprotective mechanisms for some of these natural compounds have not yet been elucidated [33, 34].
Resveratrol has been widely studied as a protective agent against many CVD. Also, resveratrol has been demonstrated to possess an inhibitory effect on CYP1B1 [35]. However, the possible beneficial effects of resveratrol as an inhibitor of CYP1B1 in case of cardiac hypertrophy have never been investigated. Therefore, in the current study, we evaluated the effects of three different concentrations of resveratrol (2, 10 and 50 μM) on cardiac CYP1B1 and its associated cardiotoxic mid-chain HETE metabolites. Using two different cell lines, we investigated whether resveratrol is able to protect against Ang II-induced cellular hypertrophy. Moreover, we characterized the effect of resveratrol on both transcriptional and translational levels of CYP1B1 and its effect on the metabolite formation of cardiotoxic mid-chain HETEs including 5-, 8-, 9-, 11-, 12- and 15-HETE.
Prior to investigating the effect of resveratrol on Ang II-induced cellular hypertrophy, we tested the effect of increasing concentrations of resveratrol on cell viability. Based on the MTT assay results, resveratrol did not have any toxic effect on cell viability following treatment for 24 h at concentrations of 2–50 µM, suggesting that the concentrations used in the current study have no cytotoxic effects. This comes in agreement with previous studies that reported the safe use of resveratrol at concentration up to 100 µM without observing toxic effects [36, 37]. Therefore, we tested the potential protective effect of resveratrol concentrations (2, 10 and 50 µM) against Ang II-induced cellular hypertrophy. Treatment of both cell lines with Ang II resulted in cellular hypertrophy as evidenced by the elevation in β-MHC/α-MHC ratio and ANP. The concentration of Ang II has been chosen based on previous work done in our lab. RL-14 cells were treated for 24 h with various concentrations of Ang II (0, 1, 2.5, 5, and 10 μM) and cellular hypertrophy markers were measured using real-time PCR. Ang II increased the expression of hypertrophic markers in a concentration-dependent manner. The maximum induction was observed at the highest concentration tested, 10 μM, and it was associated with increase in the cell surface area without affecting cell viability [38]. In another study, Ang II has been shown to induce CYP1B1 protein expression at 10 μM concentration without negatively affecting cell viability in two different cell lines [39].
The ratio of β-MHC to α-MHC is regarded as a sensitive marker of cardiac hypertrophy in several models [40,41,42]. The mechanical properties of the cardiac muscle are largely dependent on the expressed isoform of MHC [43]. Several studies have reported that α-MHC isoform has greater ATPase activity than β-MHC, and shifting from α-MHC to β-MHC isoform during cardiac hypertrophy is considered as an adaptive response for improved energy economy. However, it also leads to disruption in the contractile machinery of the heart resulting in impaired cardiac function and promoting diseases progression [44]. Furthermore, treatment of cells with Ang II caused a significant increase in ANP, a hypertrophic marker that is found to be increased at the mRNA and protein levels in human hypertrophic hearts supporting the correlation between higher ANP expression and cellular hypertrophy [45]. In the current study, low concentration of resveratrol (2 µM) was not able to significantly protect against Ang II-induced increase in β-MHC/α-MHC ratio and ANP. On the other hand, higher concentrations (10 and 50 µM) were able to reverse Ang II-mediated increase of hypertrophic markers to nearly control level, with no significant difference between the 2 concentrations. Notably, resveratrol alone groups did not have any significant effects on the mRNA level of hypertrophic markers in both cell lines, RL-14 and H9c2.
CYP1B1 is a heme-thiolate monooxygenase enzyme that is constitutively expressed in non-hepatic tissues including cardiovascular system, and is responsible for NADPH-dependent metabolism of endobiotics and xenobiotics [12]. Moreover, CYP1B1 expression was found to be induced in several experimental models of cardiac hypertrophy such as isoproterenol-induced, pressure overload-induced and Ang II-induced cardiac hypertrophy, pointing out to the role of Ang II in the progression of cardiac hypertrophy [10, 46, 47]. In the current study, while treatment of cells with Ang II did not show significant effects on mRNA expression level of CYP1B1, it significantly induced the protein expression of CYP1B1. This comes in agreement with previous study that used Ang II as an inducer of cardiac hypertrophy [17].
Several cardioprotective effects of resveratrol have been previously reported in various experimental and clinical settings. Experimentally, it attenuated pressure overload-induced cardiac fibrosis and diastolic dysfunction, it protected against chronic intermittent hypoxia-induced cardiac hypertrophy and it also restored the cardiac function and structure in case of pressure overload-induced cardiac hypertrophy [24, 25, 48]. Clinically, multiple reports provided evidences that resveratrol exerts protective effects on the cardiovascular system in patients with coronary artery diseases in the secondary prevention against myocardial infarction. Resveratrol prevented platelet aggregation, reduced LDL cholesterol level and enhanced left ventricular diastolic function [19, 49]. While several molecular mechanisms have been proposed to be responsible for the cardioprotective effects of resveratrol, this study is focusing on the effect of resveratrol on CYP1B1 and its associated cardiotoxic mid-chain HETEs. In the current study, resveratrol at concentrations of 10 and 50 µM was able to significantly decrease protein expression of CYP1B1 compared to Ang II group, in both cell lines. This comes in agreement with previous reports demonstrating the inhibitory effect of resveratrol on protein expression of CYP1B1 in several tissues [50, 51]. Although resveratrol inhibits CYP1B1 on the protein expression and activity levels, it has been found to inhibit other cytochrome P450 enzymes such as CYP1A1 and CYP1A2 with higher affinity to CYP1A1 [52]. Moreover, our lab has shown that resveratrol also inhibits the activity of human recombinant CYP4A11, CYP4F2 and CYP4F3B enzymes with higher potency on CYP4F2 [53].
Several lines of evidence previously demonstrated the role of cardiotoxic mid-chain HETEs in the pathogenesis of CVD such cardiac hypertrophy and heart failure. 15-HETE has been reported to elevate rat neonatal cardiomyocytes sensitivity to isoproterenol-induced β-adrenergic response. Moreover, 5-, 12-, and 15-HETE showed a direct inducing effect of cellular hypertrophy in RL-14 cells via MAPK- and NF-κB-dependent pathways [54, 55]. Inhibiting the formation of 5-HETE metabolite reduced Ang II-induced vascular remodeling and cardiac hypertrophy in mice proposing that inhibition of the 5-HETE formation might be considered as a new therapeutic target in the treatment of cardiac hypertrophy [56]. In the present study, treatment of cells with Ang II significantly induced the metabolite formation rate of the majority of mid-chain HETE metabolites including 5-, 8-, 12- and 15-HETE. Interestingly, treatment of cells with resveratrol at concentrations of 10 and 50 µM was able to significantly decrease the metabolite formation rate of mid-chain HETEs compared to Ang II-treated group, in both cell lines. These results are in line with previous study showing that resveratrol improved cardiac function and exercise performance in MI-induced heart failure through the inhibition of CYP1B1 and cardiotoxic HETE metabolites [16]. Moreover, our laboratory has previously shown that resveratrol was able to inhibit the formation of 20-HETE generated by incubation of AA with human liver microsomes. Resveratrol has shown concentration-dependent inhibition of the 20-HETE formation with maximum inhibition Imax of 54% and IC50 of 3 μM. These data suggest that inhibition of the pro-fibrotic 20-HETE may contribute to the beneficial and protective effects of resveratrol in case of cardiac hypertrophy [53].
In conclusion, the results of the current study showed that resveratrol, at concentrations of 10 and 50 μM, was able to protect against Ang-II-induced cellular hypertrophy as evidenced by significant inhibition of β-MHC/α-MHC ratio and ANP. While Ang II significantly induced the protein expression of CYP1B1 and increased the metabolite formation rate of its associated mid-chain HETEs, resveratrol was able to attenuate Ang II-mediated effects and caused a significant decrease of CYP1B1 protein expression and mid-chain HETEs. Our results provide the first demonstration that resveratrol protects against Ang II-induced cellular hypertrophy, at least in part, through CYP1B1/mid-chain HETEs-dependent mechanism.
References
Kavazis AN (2015) Pathological vs. physiological cardiac hypertrophy. J Physiol 593:3767
Shimizu I, Minamino T (2016) Physiological and pathological cardiac hypertrophy. J Mol Cell Cardiol 97:245–262
Metra M, Teerlink JR (2017) Heart failure. Lancet 390:1981–1995
Inamdar A, Inamdar A (2016) Heart failure: diagnosis, management and utilization. J Clin Med 5:62. https://doi.org/10.3390/jcm5070062
2016 Report on the Health of Canadians
Zhu L, Li C, Liu Q et al (2019) Molecular biomarkers in cardiac hypertrophy. J Cell Mol Med 23:1671–1677
Roman RJ (2002) P-450 metabolites of arachidonic acid in the control of cardiovascular function. Physiol Rev 82:131–185. https://doi.org/10.1152/physrev.00021.2001
Shoieb SM, El-Kadi AOS (2018) S-enantiomer of 19-hydroxyeicosatetraenoic acid preferentially protects against angiotensin II-induced cardiac hypertrophy. Drug Metab Dispos 46:1157–1168. https://doi.org/10.1124/dmd.118.082073
Gross G, Falck J, Gross E et al (2005) Cytochrome and arachidonic acid metabolites: role in myocardial ischemia/reperfusion injury revisited. Cardiovasc Res 68:18–25. https://doi.org/10.1016/j.cardiores.2005.06.007
Jennings BL, Sahan-Firat S, Estes AM et al (2010) Cytochrome P450 1B1 contributes to angiotensin II-induced hypertension and associated pathophysiology. Hypertens (Dallas, Tex, 1979) 56:667–674. https://doi.org/10.1161/HYPERTENSIONAHA.110.154518
Shoieb SM, El-Sherbeni AA, El-Kadi AOS (2019) Identification of 19-(S/R )hydroxyeicosatetraenoic acid as the first endogenous noncompetitive inhibitor of cytochrome P450 1B1 with enantioselective activity. Drug Metab Dispos 47:67–70. https://doi.org/10.1124/dmd.118.084657
Jennings BL, Montanez DE, May ME et al (2014) Cytochrome P450 1B1 contributes to increased blood pressure and cardiovascular and renal dysfunction in spontaneously hypertensive rats. Cardiovasc Drugs Ther 28:145–161. https://doi.org/10.1007/s10557-014-6510-4
Song CY, Ghafoor K, Ghafoor HU et al (2016) Cytochrome P450 1B1 contributes to the development of atherosclerosis and hypertension in apolipoprotein E-deficient mice. Hypertens 67:206–213. https://doi.org/10.1161/HYPERTENSIONAHA.115.06427
Anderson G, Mazzoccoli G (2019) Left ventricular hypertrophy: roles of mitochondria CYP1B1 and melatonergic pathways in co-ordinatingwider pathophysiology. Int J Mol Sci. https://doi.org/10.3390/ijms20164068
Maayah ZH, Althurwi HN, Abdelhamid G et al (2016) CYP1B1 inhibition attenuates doxorubicin-induced cardiotoxicity through a mid-chain HETEs-dependent mechanism. Pharmacol Res 105:28–43. https://doi.org/10.1016/j.phrs.2015.12.016
Matsumura N, Takahara S, Maayah ZH et al (2018) Resveratrol improves cardiac function and exercise performance in MI-induced heart failure through the inhibition of cardiotoxic HETE metabolites. J Mol Cell Cardiol 125:162–173. https://doi.org/10.1016/j.yjmcc.2018.10.023
Elkhatali S, Maayah ZH, El-Sherbeni AA et al (2017) Inhibition of mid-chain HETEs protects against angiotensin II-induced cardiac hypertrophy. J Cardiovasc Pharmacol 70:16–24. https://doi.org/10.1097/FJC.0000000000000494
Cione E, La Torre C, Cannataro R et al (2019) Quercetin, epigallocatechin gallate, curcumin, and resveratrol: from dietary sources to human microRNA modulation. Molecules. https://doi.org/10.3390/molecules25010063
Bonnefont-Rousselot D (2016) Resveratrol and cardiovascular diseases. Nutrients. https://doi.org/10.33390/nu8050250
Abdelgawad IY, Grant MKO, Zordoky BN (2019) Leveraging the cardio-protective and anticancer properties of resveratrol in cardio-oncology. Nutrients. https://doi.org/10.3390/nu11030627
Mikstacka R, Baer-Dubowska W, Wieczorek M, Sobiak S (2008) Thiomethylstilbenes as inhibitors of CYP1A1, CYP1A2 and CYP1B1 activities. Mol Nutr Food Res. https://doi.org/10.1002/mnfr.200700202
Beedanagari SR, Bebenek I, Bui P, Hankinson O (2009) Resveratrol inhibits dioxin-induced expression of human CYP1A1 and CYP1B1 by inhibiting recruitment of the aryl hydrocarbon receptor complex and RNA polymerase II to the regulatory regions of the corresponding genes. Toxicol Sci 110:61–67. https://doi.org/10.1093/toxsci/kfp079
Chen Z-H (2004) Resveratrol inhibits TCDD-induced expression of CYP1A1 and CYP1B1 and catechol estrogen-mediated oxidative DNA damage in cultured human mammary epithelial cells. Carcinogenesis 25:2005–2013. https://doi.org/10.1093/carcin/bgh183
Zou LX, Chen C, Yan X et al (2019) Resveratrol attenuates pressure overload-induced cardiac fibrosis and diastolic dysfunction via PTEN/AKT/Smad2/3 and NF-κB signaling pathways. Mol Nutr Food Res. https://doi.org/10.1002/mnfr.201900418
Guan P, Sun ZM, Wang N et al (2019) Resveratrol prevents chronic intermittent hypoxia-induced cardiac hypertrophy by targeting the PI3K/AKT/mTOR pathway. Life Sci. https://doi.org/10.1016/j.lfs.2019.116748
Chen C, Zou LX, Lin QY et al (2019) Resveratrol as a new inhibitor of immunoproteasome prevents PTEN degradation and attenuates cardiac hypertrophy after pressure overload. Redox Biol 20:390–401. https://doi.org/10.1016/j.redox.2018.10.021
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275
Cardiovascular diseases. https://www.who.int/health-topics/cardiovascular-diseases/#tab=tab_1. Accessed 18 Feb 2020
Ong CE, Pan Y, Mak JW (2017) The roles of cytochromes P450 in vascular biology and cardiovascular homeostasis. Int J Clin Exp Med 10:1624–1636
Elbekai RH, El-Kadi AOS (2006) Cytochrome P450 enzymes: central players in cardiovascular health and disease. Pharmacol Ther 112:564–587
Malik KU, Jennings BL, Yaghini FA et al (2012) Contribution of cytochrome P450 1B1 to hypertension and associated pathophysiology: a novel target for antihypertensive agents. Prostaglandins Other Lipid Mediat 98:69–74
Maayah ZH, El-Kadi AOS (2016) The role of mid-chain hydroxyeicosatetraenoic acids in the pathogenesis of hypertension and cardiac hypertrophy. Arch Toxicol 90:119–136
Pallazola VA, Davis DM, Whelton SP et al (2019) A clinician’s guide to healthy eating for cardiovascular disease prevention. Mayo Clin Proc Innov Qual Outcomes 3:251–267. https://doi.org/10.1016/j.mayocpiqo.2019.05.001
Hartley L, Igbinedion E, Holmes J et al (2013) Increased consumption of fruit and vegetables for the primary prevention of cardiovascular diseases. Cochrane Database Syst Rev. https://doi.org/10.1002/14651858.CD009874.pub2
MacPherson L, Matthews J (2010) Inhibition of aryl hydrocarbon receptor-dependent transcription by resveratrol or kaempferol is independent of estrogen receptor α expression in human breast cancer cells. Cancer Lett 299:119–129. https://doi.org/10.1016/j.canlet.2010.08.010
Hwang JT, Kwon DY, Park OJ, Kim MS (2008) Resveratrol protects ROS-induced cell death by activating AMPK in H9c2 cardiac muscle cells. Genes Nutr 2:323–326. https://doi.org/10.1007/s12263-007-0069-7
Chang EP, Kim MJ, Jong HL et al (2007) Resveratrol stimulates glucose transport in C2C12 myotubes by activating AMP-activated protein kinase. Exp Mol Med 39:222–229. https://doi.org/10.1038/emm.2007.25
Maayah ZH, Abdelhamid G, El-Kadi AOS (2015) Development of cellular hypertrophy by 8-hydroxyeicosatetraenoic acid in the human ventricular cardiomyocyte, RL-14 cell line, is implicated by MAPK and NF-κB. Cell Biol Toxicol 31:241–259. https://doi.org/10.1007/s10565-015-9308-7
Alammari AH, Shoieb SM, Maayah ZH, El-Kadi AOS (2020) Fluconazole represses cytochrome P450 1B1 and its associated arachidonic acid metabolites in the heart and protects against angiotensin II-induced cardiac hypertrophy. J Pharm Sci. https://doi.org/10.1016/j.xphs.2020.03.016
Qi HP, Wang Y, Zhang QH et al (2015) Activation of peroxisome proliferator-activated receptor γ (PPARγ) through NF-κB/brg1 and TGF-β 1 pathways attenuates cardiac remodeling in pressure-overloaded rat hearts. Cell Physiol Biochem 35:899–912. https://doi.org/10.1159/000369747
Wang B, Ouyang J, Xia Z (2006) Effects of triiodo-thyronine on angiotensin-induced cardiomyocyte hypertrophy: reversal of increased beta-myosin heavy chain gene expression. Can J Physiol Pharmacol 84:935–941
Chassagne C, Wisnewsky C, Schwartz K (1993) Antithetical accumulation of myosin heavy chain but not α-actin mRNA isoforms during early stages of pressure-overload-induced rat cardiac hypertrophy. Circ Res 72:857–864. https://doi.org/10.1161/01.RES.72.4.857
Alpert NR, Brosseau C, Federico A et al (2002) Molecular mechanics of mouse cardiac myosin isoforms. Am J Physiol - Hear Circ Physiol 283:H1446–H1454. https://doi.org/10.1152/ajpheart.00274.2002
Gupta MP (2007) Factors controlling cardiac myosin-isoform shift during hypertrophy and heart failure. J Mol Cell Cardiol 43:388–403
Kessler-Icekson G, Barhum Y, Schaper J et al (2002) ANP expression in the hypertensive heart. Exp Clin Cardiol 7:80–84
Maayah ZH, Althurwi HN, El-Sherbeni AA et al (2017) The role of cytochrome P450 1B1 and its associated mid-chain hydroxyeicosatetraenoic acid metabolites in the development of cardiac hypertrophy induced by isoproterenol. Mol Cell Biochem 429:151–165. https://doi.org/10.1007/s11010-017-2943-y
El-Sherbeni AA, El-Kadi AOS (2014) Alterations in cytochrome P450-derived arachidonic acid metabolism during pressure overload-induced cardiac hypertrophy. Biochem Pharmacol 87:456–466. https://doi.org/10.1016/j.bcp.2013.11.015
Dong Q, Wu Z, Li X et al (2014) Resveratrol ameliorates cardiac dysfunction induced by pressure overload in rats via structural protection and modulation of Ca2+ cycling proteins. J Transl Med 12:323. https://doi.org/10.1186/s12967-014-0323-x
Magyar K, Halmosi R, Palfi A et al (2012) Cardioprotection by resveratrol: a human clinical trial in patients with stable coronary artery disease. Clin Hemorheol Microcirc 50:179–187. https://doi.org/10.3233/CH-2011-1424
Ghosh J, Chowdhury AR, Srinivasan S et al (2018) Cigarette smoke toxins-induced mitochondrial dysfunction and pancreatitis involves aryl hydrocarbon receptor mediated Cyp1 gene expression: protective effects of resveratrol. Toxicol Sci 166:428–440. https://doi.org/10.1093/toxsci/kfy206
Huderson AC, Rekha Devi PV, Niaz MS et al (2019) Alteration of benzo(a)pyrene biotransformation by resveratrol in Apc Min/+ mouse model of colon carcinogenesis. Invest New Drugs 37:238–251. https://doi.org/10.1007/s10637-018-0622-9
Chun YJ, Kim MY, Guengerich FP (1999) Resveratrol is a selective human cytochrome P450 1A1 inhibitor. Biochem Biophys Res Commun 262:20–24. https://doi.org/10.1006/bbrc.1999.1152
El-Sherbeni AA, El-Kadi AOS (2016) Repurposing resveratrol and fluconazole to modulate human cytochrome P450-mediated arachidonic acid metabolism. Mol Pharm 13:1278–1288. https://doi.org/10.1021/acs.molpharmaceut.5b00873
Maayah ZH, El-Kadi AOS (2016) 5-, 12- and 15-Hydroxyeicosatetraenoic acids induce cellular hypertrophy in the human ventricular cardiomyocyte, RL-14 cell line, through MAPK- and NF-κB-dependent mechanism. Arch Toxicol 90:359–373. https://doi.org/10.1007/s00204-014-1419-z
Zhang L, Li Y, Chen M et al (2014) 15-LO/15-HETE mediated vascular adventitia fibrosis via p38 MAPK-dependent TGF-β. J Cell Physiol 229:245–257. https://doi.org/10.1002/jcp.24443
Revermann M, Mieth A, Popescu L et al (2011) A pirinixic acid derivative (LP105) inhibits murine 5-lipoxygenase activity and attenuates vascular remodelling in a murine model of aortic aneurysm. Br J Pharmacol 163:1721–1732. https://doi.org/10.1111/j.1476-5381.2011.01321.x
Acknowledgements
This work was supported by a grant from the Canadian Institutes of Health Research [Grant 106665] to A.O.S.E. S.M.S. is the recipient of Alberta Innovates Graduate Student Scholarship and Alberta Graduate Excellence Scholarship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shoieb, S.M., El-Kadi, A.O.S. Resveratrol attenuates angiotensin II-induced cellular hypertrophy through the inhibition of CYP1B1 and the cardiotoxic mid-chain HETE metabolites. Mol Cell Biochem 471, 165–176 (2020). https://doi.org/10.1007/s11010-020-03777-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-020-03777-9