Abstract
Aging involves complex biological processes leading to the decline of living organisms. As population lifespan increases worldwide, the importance of identifying factors underlying healthy aging has become critical. Integration of multi-modal datasets is a powerful approach for the analysis of complex biological systems, with the potential to uncover novel aging biomarkers. In this study, we leveraged publicly available epigenomic, transcriptomic and telomere length data along with histological images from the Genotype-Tissue Expression project to build tissue-specific regression models for age prediction. Using data from two tissues, lung and ovary, we aimed to compare model performance across data modalities, as well as to assess the improvement resulting from integrating multiple data types. Our results demostrate that methylation outperformed the other data modalities, with a mean absolute error of 3.36 and 4.36 in the test sets for lung and ovary, respectively. These models achieved lower error rates when compared with established state-of-the-art tissue-agnostic methylation models, emphasizing the importance of a tissue-specific approach. Additionally, this work has shown how the application of Hierarchical Image Pyramid Transformers for feature extraction significantly enhances age modeling using histological images. Finally, we evaluated the benefits of integrating multiple data modalities into a single model. Combining methylation data with other data modalities only marginally improved performance likely due to the limited number of available samples. Combining gene expression with histological features yielded more accurate age predictions compared with the individual performance of these data types. Given these results, this study shows how machine learning applications can be extended to/in multi-modal aging research. Code used is available at https://github.com/zroger49/multi_modal_age_prediction.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Aging is an intricate biological process that encompasses several cellular and molecular changes, ultimately leading to a gradual decline in tissue function and regenerative capacity. This complex phenomenon is driven by a range of mechanisms that contribute to the aging phenotype, including genomic instability, telomere attrition, mitochondrial dysfunction, altered intercellular communication and gene expression (Consortium, 2020; Rivero-Segura et al., 2020). While human lifespans have greatly increased in the past decades, the same cannot always be said for healthspans, the period when individuals are considered healthy (Beard et al., 2016). The quest to extend healthspan and enhance the quality of life for the elderly has prompted a surge of interest in developing interventions targeting the underlying mechanisms of age-related diseases (Rattan, 2018).
The most popular approach for age estimation is to train models based on diverse biological features using machine learning techniques, colloquially referred to as “aging clocks”. Currently, the most well-studied and accurate models are based on molecular data using DNA methylation. DNA methylation is vital to the developmental cycle of mammals and plays a key role in signaling and modifying the aging process (Smith & Meissner, 2013). In addition to DNA methylation, other epigenetic markers, such as histone marks, have also been associated with the aging process (Yang et al., 2010). Importantly, reversing epigenetic markers might open new venues to therapeutically target aging-related diseases (Yang et al., 2010).
Beyond epigenetic modifications, other biological data modalities are associated with the aging process, including gene expression (Viñuela et al., 2018; Melé et al., 2015), protein and metabolite abundances (Moaddel et al., 2021; Menni et al., 2013), and telomere length (Demanelis et al., 2020), demonstrating the potential for aging models leveraging different data types. Furthermore, current research has unveiled organ-specific aging signatures in human transcriptomes (Shokhirev & Johnson, 2021; Glass et al., 2013), plasma proteomes (Oh et al., 2023), and metabolic profiles (Fang et al., 2023), emphasizing the importance of modeling age in a tissue-specific manner.
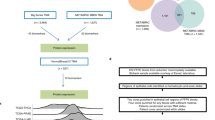
Combining different types of data, including epigenomic, proteomics, metabolomics, transcriptomic, and microbiomics, is a promising approach to unveil distinctive molecular aging patterns in individuals (Zhang, 2023). In a recent study, we tackled this issue using lung tissue and incorporated gene expression, DNA methylation, telomere length, and histological image data (Moraes et al., 2023). Here, we extend this work in several key ways: (1) we refined the previously implemented pipelines for age prediction accuracy in gene expression and methylation, by improving data preprocessing, feature selection, and adding a step to deal with data imbalance, leading to an overall improvement in performance, (2) improved the age prediction using histological images by using Hierarchical Image Pyramid Transformer (HIPT) to extract meaningful histological features, (3) evaluated the result of integrating different combinations of data modalities using a wider range of approaches, and (4) extended our prediction framework to include an additional tissue—the ovary. We selected the ovary to extend our work due to the fact that this tissue has been absent from age prediction literature (Horvath, 2013; Hannum et al., 2013; Lima Camillo et al., 2022), coupled with the importance of understanding the impact of aging in this tissue (Wang et al., 2023). Furthermore, it allows us to evaluate our prediction pipelines in both somatic and sexual tissues. A scheme of the pipeline used in this work is represented in Fig. 1.
2 Related work
DNA methylation has been widely used for age estimation and is one of the main hallmarks of the aging process (López-Otín et al., 2013). As early as 2011, Bocklandt et al. (2011) used saliva samples to build the first age estimator, achieving a MAE (mean absolute error) of 5.2 years. Two years later, two important aging models were introduced. The first, developed by Hannum et al. (2013) (colloquially named Hannum clock), was trained using 656 blood samples and achieved a MAE of 3.9 years, greatly improving on previous work. Although this model was trained on blood tissue samples, the authors were able to adjust the model to work with different tissues, producing comparable error rates. In the same year, Horvath (2013) proposed another model (Horvath clock), trained on a multi-tissue dataset of over 8,000 samples across 51 tissues. Despite age-induced methylation being highly tissue-specific (Slieker et al., 2018), the proposed model was able to generalize to most tested tissue and cell types, resulting in a MAE of 3.6 years. For both of these models, the authors used penalized regression techniques to drastically reduce the high dimensionality of the methylation arrays (\(\sim \) 450,000 for the Hannum clock and \(\sim \) 27,000 for the Horvath clock) to 71 and 353 features, respectively (Hannum et al., 2013; Horvath, 2013). More recently, neural networks (NN) have been successfully applied to epigenetic data for age prediction, achieving a MAE of 2.7, 3.8, and 2.15 years respectively (Galkin et al., 2021; Li et al., 2022; Lima Camillo et al., 2022). The latter of these models, named AltumAge, was trained with a multi-tissue dataset and demonstrated particularly promising results. Other types of data, including RNA expression data (i.e. transcriptomics), have also been used to predict biological age (Peters et al., 2015) (7.8-years MAE). This model was applied with the Horvath and the Hannum clock to a set of common samples and uncovered that the error rates of each data modality were correlated with different aging-related phenotypes. This suggests that gene expression and methylation may provide complementary information for age prediction. A number of age models have been built using other types of molecular data, including protein abundances (Sayed et al., 2021; Kristic et al., 2014; Tanaka et al., 2018; Lehallier et al., 2020), metabolites quantities (Akker et al., 2020; Robinson et al., 2020; Hertel et al., 2016), taxonomic profiling of gut microbiota (Galkin et al., 2020; Chen et al., 2022), blood biomarkers (Mamoshina et al., 2018), and chromatin accessibility (Morandini et al., 2024). Despite the large number of data modalities found in the literature, there has been a very limited number of attempts at generating aging models using multi-modal datasets. We previously focused on this problem and attempted to combine gene expression, DNA methylation, and histological images for age prediction using an ensemble approach (Moraes et al., 2023). However, this approach led to very similar results when compared with individual data modalities. As far as we know, the only other example of this class of models is Precious1GPT, which combines methylation and gene expression data (Urban et al., 2023), where the performance of methylation models alone was better than the combined dataset (methylation data MAE 4.23, gene expression data MAE 6.28, combined MAE 5.62).
3 Data description and preparation
All the data used in this study was generated in the context of the GTEx (Genotype-Tissue Expression) project (GTEx, 2020) and is publicly available online. We focused on lung and ovary tissue across four data modalities explored: gene expression data, DNA methylation data, telomere length data, and histological images data (Fig. 2a, b).
DNA methylation data generated in Oliva et al. (2023) was downloaded from the Gene expression Omnibus (GEO; accession number GSE213478). The processed dataset contains roughly 750,000 features corresponding to CpG sites, measured in \(\beta \)-values, with values ranging from 0 to 1. The \(\beta \)-values were converted to M-values. In contrast to M-values, \(\beta \)-values have been shown to suffer from heteroscedasticity in highly methylated and unmethylated positions (values near 1 and 0 respectively) (Du et al., 2010).
Gene expression data (both raw counts and normalized TPM values) generated using RNA-seq was downloaded from the GTEx data portal (https://www.gtexportal.org/home/). For each tissue, we selected protein-coding and long non-coding RNAs with minimum TPM of 1 in at least 20% of samples (15,746 features in lung; 14,712 features in ovary). Following filtering, the selected gene features underwent a log2(x+1) transformation.
Whole slide images (WSI) were downloaded from the GTEx Histological Image Viewer (https://gtexportal.org/home/histologyPage) and divided into smaller patches of \([4096 \times 4096]\) pixels (with 35 patches per WSI on average). Then, for each patch k, we extracted two types of features, \([CLS]_{4096}^{(k)}\) and \([CLS]_{256}^{(j)}\) (where j is the index of the j-th patch of \([256 \times 256]\) within the k-th patch of \([4096 \times 4096]\), and [CLS] is the classifier token), utilizing the HIPT architecture. We further aggregated and combined information from these two features sets, culminating into 3 types of features \([CLS]_{4096}^{(WSI)}\), \([CLS]_{256}^{(WSI)}\) and \([CLS]_{4096,256}^{(WSI)}\).
Telomere length (TL) data generated in Demanelis et al. (2020) was downloaded from the GTEx portal. This data type is represented by a single feature, relative telomere length, which measures telomere repeat abundance in a DNA sample relative to a standard sample.
Summary of the pipeline implemented for model training. Data was retrieved from the GTEx portal and repositories containing data generated for the companion papers. We split the data from each tissue and each modality into train-test sets, with the test samples being common to all data modalities within a tissue. Model optimization was performed using fivefold cross-validation. Feature selection was implemented to decrease the feature space in gene expression and DNA methylation. Histological features were extracted from images using the HIPT architecture. To avoid data leakage, feature selection was performed independently for each fold. Model training was performed using a variety of algorithms. The final model evaluation was performed on the common test set
4 Overview of prediction model methodology
For each tissue, we split all data modalities into train-test sets. We verified that age labels were skewed to older individuals (Fig. 2c, d). Consequently, during the train-test split, we ensured the representation of younger individuals in the dataset. Specifically, we performed a stratified partitioning of the methylation dataset with an 80-20 overall split and a 50-50 split for younger ages (< 45 years old). This ensured that younger individuals were represented in the test dataset, allowing us to evaluate the ability of the trained models to generalize in this age range. Test samples were common to all data modalities. For the remaining data modalities, samples not included in the common test set were used for model training.
Number of samples in each data modality in lung (a) and ovary data (b). The overlap between data type was not complete. Common samples represent the number of samples in common across the 4 data types. Distribution of age in the train set of each data modality in lung (c) and ovary (d) (Color figure online)
State-of-the-art age prediction models were applied to the epigenetic data, in order to establish a baseline for model comparison. These models are based on either selected methylation features (Hannum et al., 2013; Horvath, 2013) or all the features contained in the Illumina Infinium HumanMethylation27 array (Lima Camillo et al., 2022). A percentage of these features was missing from our processed epigenetic dataset. Before applying each model to our data, we imputed the corresponding missing features, by taking the mean of \(\beta \)-values of the all the features in each sample. We note that this processing step was only performed in the context of age prediction using these models, and were not considered during our model training pipeline. After data imputation, each methylation model was applied to the train-test set.
The ML pipelines were optimized using the optuna package (Akiba et al., 2019) in a fivefold cross-validation strategy (CV). Briefly, we divided the train dataset into 5 partitions (folds). We then trained the model on fourfolds (training folds), while the remaining fold was held out for validation. We repeated this process 5 times, with each fold serving as validation one time. All steps from the pipeline, including feature selection and Label Distribution Smoothing (LDS) were performed using only the training-folds to avoid data leakage. The best combination of parameters were selected based on average mean absolute error (MAE). These parameters were then applied to the whole train data to train the final model. Alternatively nested CV could had been applied, completely separating the tasks for hyper-parameter tuning and performance evaluation.
Due to the high dimensionality of the molecular datasets (gene expression and methylation), we evaluated several feature selection methods in order to decrease the feature space (Section A.5). In particular we identified methylation and gene expression features that significantly change with age in each tissue, by performing differential expression analysis using limma (Ritchie et al., 2015). Briefly, this method evaluates change in expression/methylation, by fitting feature-wise linear models using the target variable (in this case age). This frameworks allows for the inclusion of other covariates, in order to correct for potentially confounding effects. As such, we also included both technical (e.g. ischemic time and RIN) and biological (sex and BMI) covariates. Elastic net served as an additional feature selection method, and was considered before the training of non-linear models.
Elastic Net (EN) and Gradient Boosting Trees (GBT) algorithms were used to build the predictive models for methylation and gene expression, using their implementation in the sklearn and lightgbm (Ke et al., 2017) python packages respectively. This allowed us to test linear and non-linear algorithms for age prediction using molecular data. Furthermore, both elastic net and GBT are computationally efficient, easy to interpret, and have previously shown good results for prediction tasks using molecular data (Takahashi et al., 2020; Horvath, 2013; Hannum et al., 2013).
A Gaussian quantile transformation was applied prior to model training to reduce the right-skewed distribution. This process spread out the values, particularly those centered around 0, and decreased the influence of outliers. To further address age skewness, we implemented sample re-weighting using LDS (Yang et al., 2021).
Regarding the histological data models, for each Whole Slide Image (WSI) we extracted features using the pre-trained HIPT model (Chen et al., 2023). Similar to molecular datasets, we used EN for age prediction. Furthermore, we also implemented a custom-designed Multi-Layer Perceptron (MLP) that served as an encoder, forcing the dimensionality of its input data to be progressively reduced throughout the forward propagation process, followed by a linear layer for age prediction.
Similarly to the molecular datasets, we applied the LDS technique to address age skewness. We assessed TL, measured using Luminex-based methods (Demanelis et al., 2020), as a predictive biomarker of age using a multiple linear regression model including this feature and other demographic variables of the donors.
In the context of multi-modal modeling, we systematically explored various permutations of data modalities and performed integration using EN, GBT, and MLP. In order to ensure comparability across different combinations of data types, we constrained model training to the samples encompassed by all four modalities (Sect. 5.4) or shared samples between gene expression and histology (Sect. 5.5). Furthermore, we repeated the training process for each data type using only the shared samples in order to assess the benefits of multi-modal data integration.
We evaluated the performance between data modalities and combinations of modalities using Nadeau and Bengio’s variance-corrected t-test by comparing the tested model to the corresponding baseline on the MAE across the fivefolds.
A more detailed overview of our framework can be found in appendix A (Supplementary Methods). The code used in this work can be found in https://github.com/zroger49/multi_modal_age_prediction.
5 Experimental results
5.1 State-of-the-art methylation clocks performance
In order to establish the current state-of-the-art performance and a baseline for all future model comparisons, we tested the performance of three epigenetic-based models on our data: the Horvarth clock (Horvath, 2013), the Hannum clock (Hannum et al., 2013) and AltumAge (Lima Camillo et al., 2022). Each model was applied independently to the train-test sets (Table 1, Supplementary table 1). The performance obtained for the Horvath clock and AltumAge in lung data (train set MAE = 4.39 and 4.07 respectively) was within 1 year of the reported accuracy for these tissues (Horvath, 2013; Lima Camillo et al., 2022). The Hannum clock reported the largest error (train set MAE 7.23). Although lung tissue was used to validate this model (Hannum et al., 2013), the authors did not report tissue-specific error rates. Applying the same models to ovary data yielded a MAE >10 years. Although both Horvarth clock and AltumAge have been reported to yield similar error rates in certain tissues (Horvath, 2013; Lima Camillo et al., 2022), ovary data was not present in either their train or validation datasets, and therefore a direct comparison is not possible. We further applied these models to two additional tissues: colon and prostate (Supplementary Table 1). Analyzing all four tissues we observed that, for our data:
-
The Hannum clock yields large errors (MAE >10 years) for all tissues except lung, where it still performs worse than the other two models.
-
Overall, AltumAge has lower MAE compared with the Horvarth clock.
-
All molecular clocks yield large error rates (MAE >10 years) in the ovary (with the exception of AltumAge in the test set; MAE = 8.74).
The poor performance of the Hannum clock might be due to the high percentage of missing features in the processed methylation dataset (9 out of 71–13%). The Horvarth clock and AltumAge also contained missing features, although to a lesser extent (24/354–6.7% and 524/20,318–2.6%). Indeed, in order to apply these models to our data, we had to perform a data imputation step (Section A.2), which might affect age estimation, since the imputed values might not completely reflect reality.
With the results delineated above, we selected AltumAge as the baseline for comparison with our methylation models.
5.2 Model performance within individual data modalities
Performance of single data modality models on the test set. Panels a–d represent the performance in lung data in methylation (a), gene expression (b), histological images (c) and telomeres (d), while E–H represents the performance of ovary in methylation (e), gene expression (f), histological images (g) and telomeres (h) data. Mean absolute error—mae, rmse—root mean squared error, med—Median absolute error, cor—correlation, R2—coefficient of determination (Color figure online)
We developed tissue-specific aging models for each data type using several machine learning algorithms, including linear (EN) and non-linear (GBT and NN) algorithms. The performance of the best model is summarized in Table 1 and Fig. 3a–d for lung, e–f for ovary. EN models were found to be the most effective for both methylation and gene expression models, while neural networks were found to be more effective for modeling histological images. A more complete version of this table, along with the performance of other tested approaches is presented in Supplementary table 2.
To ensure fairness in performance comparison across data modalities, we defined a common test set across the four types of data (Fig. 1). Despite being the data modality with the lowest sample size, methylation models achieved the lowest error rates both during CV and in the test set across both tissues (lung test MAE 3.36, Fig. 3a–d; ovary test MAE 4.36, Fig. 3e–h).
In order to compare performance of our developed models to the state-of-the-art approach, we derived the fivefold performance of AltumAge, by dividing the predicted age into 5 sets, following the same splits used for CV during model training. The lung model performed significantly better than AltumAge model (p value < 0.05). The ovary methylation model also performed significantly better than AltumAge in this tissue, but this comparison holds less meaning, the multi-tissue methylation clocks did not generalize in this tissue (Supplementary Table 1). The improvement verified across models can be attributed to the fact that AltumAge model was trained on a multi-tissue dataset (Lima Camillo et al., 2022), and therefore is not as accurate in capturing the tissue-specific methylation patterns observed in lung and ovary (Christensen et al., 2009).
Gene expression and histological images have very similar performances, with MAE 5–6 years for gene expression and 5–7 years for histology. The error rates obtained for gene expression are within previously reported results (Peters et al., 2015; Fleischer et al., 2018; Mamoshina et al., 2018; Xia et al., 2020; Holzscheck et al., 2021; Meyer & Schumacher, 2021). In contrast, for histological images, we observed a major improvement in performance compared with our previous work (Moraes et al., 2023) performed in the same dataset (lung test MAE 5.37 vs 8.15; lung test R2 0.69 vs 0.35). Finally, telomere length yielded the worst models (CV MAE \(\sim \) 9 year, test MAE >10 years). Nevertheless, TL showed significant negative correlation with age in lung even after correcting for demographic variables (p value < 0.05), in line with previous results (Demanelis et al., 2020).
5.3 Model interpretation
SHAP values for DNA methylation and gene expression age predictors. Results are shown for the a lung methylation model, b ovary methylation model, c lung gene expression model and d ovary methylation model. SHAP values were computed based on the test set. Only the top 10 features (mean absolute SHAP) in each model are represented. In methylation models, the features are represented by the CpG site identifier and the nearest gene. Dots are colored according to their scaled \(\beta \)-values (methylation models) or TPM values (gene expression), with red dots representing higher values and blue dots representing lower values. The position in the X-axis corresponds to negative (decrease in age) or positive (increase in age) feature impact (Color figure online)
To identify and rank the most important features in methylation and gene expression models for each tissue, we employed SHapley Additive exPlanations (SHAP) (Lundberg & Lee, 2017). We identified several features that have been previously described to be related with aging in human tissues. For example, in the lung methylation model the most important feature, cg27320127 (Fig. 4a) is one of the predictive features in at least four previously described epigenetic age models (Jones et al., 2015). Also in the lung model, cg00590036, the third most important feature, was found to be altered with age in adipose tissue (Rönn et al., 2015). In ovary, cg05708550 a site near the KDM3B gene was the fifth most important feature, with higher methylation levels leading to higher age predictions (Fig. 4b). Despite no studies associating this site directly with aging, decrease in KDM3B expression has been correlated with retinal disruption (An et al., 2022).
In gene expression, ZNF518B, the most important gene in the lung model, has been linked to human longevity and is negatively correlated with age (Bou Sleiman et al., 2020), consistent with finding in this work (Fig. 4c). In contrast, the expression for the gene EDA2R has been found to be positively correlated with age in several human tissues, including lung (Jeong et al., 2020; Melé et al., 2015). In ovary tissue, the most important gene was TRIM59 (Fig. 4d), which was negatively correlated with age. Interestingly, previous epigenetic studies have identified methylation markers related to aging in this gene (Jung et al., 2019; Wezyk et al., 2018).
In summary, our findings indicate that our models effectively capture molecular age-related features in human tissues, unveiling potential aging biomarkers, some of which have not been previously reported in the literature.
5.4 Combining methylation with other data modalities has a limited effect on model performance
Despite having fewer samples, methylation data outperformed the other three data types in age prediction across both tissues in the test set (Table 1). We set out to investigate if integrating other data modalities with methylation would improve model performance. We trained several models with different data combinations: methylation + gene expression, methylation + histology, methylation + gene expression + histology and methylation + gene expression + histology + telomere length. To ensure a fair comparison, we trained these models using only the common samples across the four data modalities. Furthermore, as a baseline, we retrained the methylation model using the same subset of samples.
First, we evaluated the performance using EN, since this was the method that performed best for both methylation and gene expression. Combining methylation with gene expression and histology led to the best results in both tissues in the cross-validation with a marginal and non-significant difference (3.75 ± 0.56 vs 3.80 ± 0.5 MAE in lung, p value = 0.36; 4.17±0.46 vs 4.35±0.33 MAE in ovary, p value = 0.29). All the models also showed a similar performance in the test set, with the model including methylation and histology having slightly lower MAE.
We obtained similar results when integrating the data using GBT and NN (Supplementary table 3). In the former, although the performance was better in CV when compared with EN, the MAE in the test set was higher (+0.5-1 year). Employing NN, we observed enhanced performance by combining methylation with gene expression and histological images compared to integrating methylation with each data type individually (p value < 0.05) in ovary tissue, while in lung tissue, integrating these three data modalities was equivalent to combining methylation with gene expression (p value > 0.05). Neverthelss, the performance of the combined models was similar to the downsampled EN methylation models (p value > 0.05).
SHAP analysis (Lundberg & Lee, 2017) was used in the EN models to quantify the contribution of each data type. Methylation was the most significant data type, with a total feature contribution ranging from 81.14% to 98.65% (Fig. 5). It should be noted that methylation also has the highest number of features per model. Telomere length did not contribute to model prediction. We analyzed the SHAP values of the model integrating methylation, gene expression and histology to get a sense of the most important features when integrating these data modalities. In lung we recapitulate several features, such as cg27320127 and cg00590036. Among the genes in the top 15 features we found H4C12 (H4 Clustered Histone 12), of particular interest, since the depletion of histones has been shown to occur during human aging (Dubey et al., 2024). In ovary we found that a long non-coding RNA (EPHA5-AS1) gene had the highest SHAP values, with an increasing expression with age. No previous study has linked this gene with aging.
In summary, our results demonstrate that, for these tissues, integrating methylation data with histological and gene expression data leads to only marginal improvements in model performance.
SHAP analysis for the multi-modal models using methylation data. a Percentage of contribution of each data type for the final model prediction (methyl—DNA methylation, Gexp—gene expression, Hist—histological features and TL—telomere length). b, c SHAP values for the model including methylation, gene expression and histological images in (B) lung and (C) ovary. SHAP values were computed based on the test set. Only the top 15 features (based on mean absolute SHAP) in each model are represented. Methylation features are represented by the CpG site identifier and the nearest gene. Red dots represent higher values and blue dots represent lower values. The position in the X-axis corresponds to negative (decrease in age) or positive (increase in age) feature impact (Color figure online)
5.5 Combining gene expression with histology improves their individual model performance
The small number of samples available could be hampering performance improvements in the multi-modal models. As methylation is one of the main hallmarks of aging (López-Otín et al., 2023), it is expected to require less samples to accurately model age compared with histology and gene expression.
We performed a second integration experiment by combining histology and gene expression, and comparing the performance of these multi-modal models with their single data type variant (Table 3, Supplementary Table 4). Similar to the previous section, we trained individual gene expression and histological models using only the common samples between these two data modalities. The EN model combining gene expression and histology performed significantly better in lung compared with the model using only gene expression (p value = 0.005) and histology (p value = \(2.363 \times 10^{-9}\)). In ovary, we verified a similar tendency. In both cases, we also verified an improvement in the test set.
6 Discussion
In this work, we have leveraged four data types from the GTEx dataset to build age prediction models in two human tissues: lung and ovary (Fig. 1). Methylation and gene expression models performed in line with previous studies (Peters et al., 2015; Fleischer et al., 2018; Mamoshina et al., 2018; Xia et al., 2020; Holzscheck et al., 2021; Meyer & Schumacher, 2021; Horvath, 2013; Hannum et al., 2013; Lima Camillo et al., 2022). As expected, methylation models were the best performing despite having the lowest sample size, indicating the presence of a stronger signal for this data type.
The success of histological images for age prediction has been limited and requires further exploration. In a recent work, we explored this idea with lung data (Moraes et al., 2023). We hypothesized that a contributing factor to the limited performance of the histological models was the loss of contextual information caused by the selection of [256 \(\times \) 256] pixel patches, which focused solely on cellular-level features, therefore potentially overlooking broader tissue structures and contextual cues that might be useful for an accurate determination of age. To get around this problem, Chen et al. (2023) proposed a Visual Transformer (ViT)-based architecture, HIPT, to extract meaningful features from WSI considering three main resolutions: [16 \(\times \) 16] (cellular features), [256 \(\times \) 256] (cellular organization), and [4096 \(\times \) 4096] (tissue phenotypes). In this study, we applied this technique and developed an age model based on histological images which significantly improves on the previous work (test MAE from 8.15 to 6.29; test R2 from 0.35 to 0.59). Furthermore, we extended our framework beyond a single tissue and applied the same techniques in ovary.
Integrating methylation with other data types yielded limited improvement in predictive performance (Table 3). This may result from the low sample size of the common samples, with methylation benefiting from a stronger association with aging. We made a second experiment, focusing only on histology and gene expression data, since these modalities shared the highest number of common samples. We observed that combining these two data types significantly improved age prediction compared to their individual performances, thereby demonstrating the potential of integrating different data types in age prediction models.
This work highlighted several challenges in working with biological multi-modal datasets. First, the molecular data has more features than samples, with roughly 750,000 features by less than 200 samples in methylation and around 20,000 features by 128 to 527 samples in gene expression. Feature selection was an important step for significantly reducing the number of input features while maintaining or even contributing to more competitive model performance. Second, the datasets are skewed towards older individuals, with methylation being the most affected due to its smaller sample size. To address this issue, we implemented sample re-weighing with label distribution smoothing (Yang et al., 2021). Moreover, we stratified ages during cross-validation and the train-test split, a method demonstrated to enable fairer comparisons among individuals within similar age ranges (Li et al., 2023).
Another limitation of the analysis conducted herein is that the GTEx samples are obtained post-mortem, and the data might be skewed by a number of confounding factors, including ischemic time and cause of death that might alter the molecular fingerprint. We attempted to account for confounding factors during feature selection through differential analysis, where we explicitly corrected for ischemic time and hardy scale (a measure to categorize the severity of injuries sustained in violent deaths), as well as a number of other covariates. Despite this, the applicability of the developed models and methodology requires further evaluation and investigation.
The integration of multiple data types from the same cohort remains largely unexplored and therefore constitutes a strong point of our work. To date, there have been relatively few instances of aging research utilizing multi-modals models in the literature (Urban et al., 2023; Solovev et al., 2020). Our work pioneers the integration of transcriptomics, epigenomics, telomere data, and histological images from the same population, offering novel insights into aging research.
Data and material availability
Age data for GTEx individuals is under protected data, and can be accessed in dbGap under the accession number phs000424.v8. (https://gtexportal.org/home/protectedDataAccess). Expression, telomere and histology data is publicly available through the GTEx Portal as downloadable files (https://www.gtexportal.org). DNA methylation data is available in GEO under the accession number GSE213478.
Code Availability
The code used in this work can be found in https://github.com/zroger49/multi_modal_age_prediction
References
Akiba, T., Sano, S., Yanase, T., Ohta, T., & Koyama, M. (2019). Optuna: A next-generation hyperparameter optimization framework. In Proceedings of the 25th ACM SIGKDD international conference on knowledge discovery and data mining.
Akker, E. B., Trompet, S., Barkey Wolf, J. J. H., Beekman, M., Suchiman, H. E. D., Deelen, J., Asselbergs, F. W., Boersma, E., Cats, D., Elders, P. M., Geleijnse, J. M., Ikram, M. A., Kloppenburg, M., Mei, H., Meulenbelt, I., Mooijaart, S. P., Nelissen, R., Netea, M. G., Penninx, B., … Slagboom, P. E. (2020). Metabolic age based on the bbmri-nl (1)h-nmr metabolomics repository as biomarker of age-related disease. Circulation: Genomic and Precision Medicine, 13(5), 541–547. https://doi.org/10.1161/CIRCGEN.119.002610
An, M.-J., Kim, J.-Y., Park, J., Kim, J., Kim, D.-H., Shin, G.-S., Lee, H.-M., Jo, A.-R., Kim, C.-H., & Kim, M. J.: Site-specific gains and losses of heterochromatin accelerate the age-related neurodegeneration through the cascading destruction of kdm3b-centered epigenomic network (2022)
Beard, J. R., Officer, A., Carvalho, I. A., Sadana, R., Pot, A. M., Michel, J. P., Lloyd-Sherlock, P., Epping-Jordan, J. E., Peeters, G., Mahanani, W. R., Thiyagarajan, J. A., & Chatterji, S. (2016). The world report on ageing and health: A policy framework for healthy ageing. Lancet, 387(10033), 2145–2154. https://doi.org/10.1016/s0140-6736(15)00516-4
Bocklandt, S., Lin, W., Sehl, M. E., Sanchez, F. J., Sinsheimer, J. S., Horvath, S., & Vilain, E. (2011). Epigenetic predictor of age. PLoS One, 6(6), 14821. https://doi.org/10.1371/journal.pone.0014821
Bou Sleiman, M., Jha, P., Houtkooper, R., Williams, R. W., Wang, X., & Auwerx, J. (2020). The gene-regulatory footprint of aging highlights conserved central regulators. Cell Reports, 32(13), 108203. https://doi.org/10.1016/j.celrep.2020.108203
Chen, R. J., Chen, C., Li, Y., Chen, T. Y., Trister, A. D., Krishnan, R. G., & Mahmood, F. (2023). Scaling vision transformers to gigapixel images via hierarchical self-supervised learning. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition (pp. 16144–16155).
Chen, Y., Wang, H., Lu, W., Wu, T., Yuan, W., Zhu, J., Lee, Y. K., Zhao, J., Zhang, H., & Chen, W. (2022). Human gut microbiome aging clocks based on taxonomic and functional signatures through multi-view learning. Gut Microbes, 14(1), 2025016. https://doi.org/10.1080/19490976.2021.2025016
Christensen, B. C., Houseman, E. A., Marsit, C. J., Zheng, S., Wrensch, M. R., Wiemels, J. L., Nelson, H. H., Karagas, M. R., Padbury, J. F., Bueno, R., Sugarbaker, D. J., Yeh, R. F., Wiencke, J. K., & Kelsey, K. T. (2009). Aging and environmental exposures alter tissue-specific dna methylation dependent upon cpg island context. PLoS Genetics, 5(8), 1000602. https://doi.org/10.1371/journal.pgen.1000602
Consortium A.A. (2020). Aging Atlas: A multi-omics database for aging biology. Nucleic Acids Research,49(D1), 825–830.
Demanelis, K., Jasmine, F., Chen, L. S., Chernoff, M., Tong, L., Delgado, D., Zhang, C., Shinkle, J., Sabarinathan, M., Lin, H., Ramirez, E., Oliva, M., Kim-Hellmuth, S., Stranger, B. E., Lai, T. P., Aviv, A., Ardlie, K. G., Aguet, F., Ahsan, H., … Pierce, B. L. (2020). Determinants of telomere length across human tissues. Science. https://doi.org/10.1126/science.aaz6876
Dubey, S. K., Dubey, R., & Kleinman, M. E. (2024). Unraveling histone loss in aging and senescence. Cells. https://doi.org/10.3390/cells13040320
Du, P., Zhang, X., Huang, C.-C., Jafari, N., Kibbe, W. A., Hou, L., & Lin, S. M. (2010). Comparison of beta-value and m-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics, 11(1), 587. https://doi.org/10.1186/1471-2105-11-587
Fang, W., Chen, S., Jin, X., Liu, S., Cao, X., & Liu, B. (2023). Metabolomics in aging research: Aging markers from organs. Frontiers in Cell and Developmental Biology. https://doi.org/10.3389/fcell.2023.1198794
Fleischer, J. G., Schulte, R., Tsai, H. H., Tyagi, S., Ibarra, A., Shokhirev, M. N., Huang, L., Hetzer, M. W., & Navlakha, S. (2018). Predicting age from the transcriptome of human dermal fibroblasts. Genome Biology, 19(1), 221. https://doi.org/10.1186/s13059-018-1599-6
Galkin, F., Mamoshina, P., Aliper, A., Putin, E., Moskalev, V., Gladyshev, V. N., & Zhavoronkov, A. (2020). Human gut microbiome aging clock based on taxonomic profiling and deep learning. iScience, 23(6), 101199. https://doi.org/10.1016/j.isci.2020.101199
Galkin, F., Mamoshina, P., Kochetov, K., Sidorenko, D., & Zhavoronkov, A. (2021). Deepmage: A methylation aging clock developed with deep learning. Aging and Disease, 12(5), 1252–1262. https://doi.org/10.14336/AD.2020.1202
Glass, D., Viñuela, A., Davies, M.N., Ramasamy, A., Parts, L., Knowles, D., Brown, A. A., Hedman, Ä. K., Small, K. S., Buil, A., Grundberg, E., Nica, A. C., Di Meglio, P., Nestle, F. O., Ryten, M., Durbin, R., McCarthy, M. I., Deloukas, P., Dermitzakis, E. T., Weale, M. E., Bataille, V., Spector, T. D., the, U.K.B.E.c., & Mu, T. C. (2013). Gene expression changes with age in skin, adipose tissue, blood and brain. Genome Biology 14(7), 75.
GTEx: The gtex consortium atlas of genetic regulatory effects across human tissues. Science 369(6509), 1318–1330 (2020)
Hannum, G., Guinney, J., Zhao, L., Zhang, L., Hughes, G., Sadda, S., Klotzle, B., Bibikova, M., Fan, J. B., Gao, Y., Deconde, R., Chen, M., Rajapakse, I., Friend, S., Ideker, T., & Zhang, K. (2013). Genome-wide methylation profiles reveal quantitative views of human aging rates. Molecular Cell, 49(2), 359–367. https://doi.org/10.1016/j.molcel.2012.10.016
Hertel, J., Friedrich, N., Wittfeld, K., Pietzner, M., Budde, K., Auwera, S., Lohmann, T., Teumer, A., Völzke, H., Nauck, M., & Grabe, H. J. (2016). Measuring biological age via metabonomics: The metabolic age score. Journal of Proteome Research, 15(2), 400–10. https://doi.org/10.1021/acs.jproteome.5b00561
Holzscheck, N., Falckenhayn, C., Sohle, J., Kristof, B., Siegner, R., Werner, A., Schossow, J., Jurgens, C., Volzke, H., Wenck, H., Winnefeld, M., Gronniger, E., & Kaderali, L. (2021). Modeling transcriptomic age using knowledge-primed artificial neural networks. NPJ Aging and Mechanisms of Disease, 7(1), 15. https://doi.org/10.1038/s41514-021-00068-5
Horvath, S. (2013). Dna methylation age of human tissues and cell types. Genome Biology, 14(10), 3156. https://doi.org/10.1186/gb-2013-14-10-r115
Jeong, I., Lim, J. H., Park, J. S., & Oh, Y. M. (2020). Aging-related changes in the gene expression profile of human lungs. Aging (Albany NY), 12(21), 21391–21403.
Jones, M. J., Goodman, S. J., & Kobor, M. S. (2015). Dna methylation and healthy human aging. Aging Cell, 14(6), 924–932. https://doi.org/10.1111/acel.12349
Jung, S. E., Lim, S. M., Hong, S. R., Lee, E. H., Shin, K. J., & Lee, H. Y. (2019). Dna methylation of the elovl2, fhl2, klf14, c1orf132/mir29b2c, and trim59 genes for age prediction from blood, saliva, and buccal swab samples. Forensic Science International: Genetics, 38, 1–8. https://doi.org/10.1016/j.fsigen.2018.09.010
Ke, G., Meng, Q., Finley, T., Wang, T., Chen, W., Ma, W., Ye, Q., & Liu, T.-Y. (2017). Lightgbm: A highly efficient gradient boosting decision tree. In Advances in neural information processing systems (Vol. 30).
Kristic, J., Vuckovic, F., Menni, C., Klaric, L., Keser, T., Beceheli, I., Pucic-Bakovic, M., Novokmet, M., Mangino, M., Thaqi, K., Rudan, P., Novokmet, N., Sarac, J., Missoni, S., Kolcic, I., Polasek, O., Rudan, I., Campbell, H., Hayward, C., … Lauc, G. (2014). Glycans are a novel biomarker of chronological and biological ages. Journals of Gerontology Series A: Biological Sciences and Medical Sciences, 69(7), 779–89. https://doi.org/10.1093/gerona/glt190
Lehallier, B., Shokhirev, M. N., Wyss-Coray, T., & Johnson, A. A. (2020). Data mining of human plasma proteins generates a multitude of highly predictive aging clocks that reflect different aspects of aging. Aging Cell, 19(11), 13256. https://doi.org/10.1111/acel.13256
Lima Camillo, L. P., Lapierre, L. R., & Singh, R. (2022). A pan-tissue dna-methylation epigenetic clock based on deep learning. NPJ Aging. https://doi.org/10.1038/s41514-022-00085-y
Li, L., Zhang, C., Liu, S., Guan, H., & Zhang, Y. (2022). Age prediction by dna methylation in neural networks. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 19(3), 1393–1402. https://doi.org/10.1109/TCBB.2021.3084596
Li, Z., Zhang, W., Duan, Y., Niu, Y., Chen, Y., Liu, X., Dong, Z., Zheng, Y., Chen, X., Feng, Z., Wang, Y., Zhao, D., Sun, X., Cai, G., Jiang, H., & Chen, X. (2023). Progress in biological age research. Frontiers in Public Health, 11, 1074274. https://doi.org/10.3389/fpubh.2023.1074274
López-Otín, C., Blasco, M. A., Partridge, L., Serrano, M., & Kroemer, G. (2013). The hallmarks of aging. Cell, 153(6), 1194–217. https://doi.org/10.1016/j.cell.2013.05.039
López-Otín, C., Blasco, M. A., Partridge, L., Serrano, M., & Kroemer, G. (2023). Hallmarks of aging: An expanding universe. Cell, 186(2), 243–278. https://doi.org/10.1016/j.cell.2022.11.001
Lundberg, S. M., Lee, S.-I. (2017). A unified approach to interpreting model predictions. In Advances in neural information processing systems (Vol. 30).
Mamoshina, P., Kochetov, K., Putin, E., Cortese, F., Aliper, A., Lee, W. S., Ahn, S. M., Uhn, L., Skjodt, N., Kovalchuk, O., Scheibye-Knudsen, M., & Zhavoronkov, A. (2018). Population specific biomarkers of human aging: A big data study using south Korean, Canadian, and Eastern European patient populations. Journals of Gerontology Series A: Biological Sciences and Medical Sciences, 73(11), 1482–1490. https://doi.org/10.1093/gerona/gly005
Melé, M., Ferreira, P. G., Reverter, F., DeLuca, D. S., Monlong, J., Sammeth, M., Young, T. R., Goldmann, J. M., Pervouchine, D. D., Sullivan, T. J., Johnson, R., Segrè, A. V., Djebali, S., Niarchou, A., Wright, F. A., Lappalainen, T., Calvo, M., Getz, G., Dermitzakis, E. T., … Guigó, R. (2015). Human genomics the human transcriptome across tissues and individuals. Science, 348(6235), 660–665. https://doi.org/10.1126/science.aaa0355
Menni, C., Kastenmuller, G., Petersen, A. K., Bell, J. T., Psatha, M., Tsai, P. C., Gieger, C., Schulz, H., Erte, I., John, S., Brosnan, M. J., Wilson, S. G., Tsaprouni, L., Lim, E. M., Stuckey, B., Deloukas, P., Mohney, R., Suhre, K., Spector, T. D., & Valdes, A. M. (2013). Metabolomic markers reveal novel pathways of ageing and early development in human populations. International Journal of Epidemiology, 42(4), 1111–9. https://doi.org/10.1093/ije/dyt094
Meyer, D. H., & Schumacher, B. (2021). Bit age: A transcriptome-based aging clock near the theoretical limit of accuracy. Aging Cell, 20(3), 13320. https://doi.org/10.1111/acel.13320
Moaddel, R., Ubaida-Mohien, C., Tanaka, T., Lyashkov, A., Basisty, N., Schilling, B., Semba, R. D., Franceschi, C., Gorospe, M., & Ferrucci, L. (2021). Proteomics in aging research: A roadmap to clinical, translational research. Aging Cell, 20(4), 13325. https://doi.org/10.1111/acel.13325
Moraes, A., Moreno, M., Ribeiro, R., & Ferreira, P. G. (2023). Predicting age from human lung tissue through multi-modal data integration. In Discovery science (pp. 644–658).
Morandini, F., Rechsteiner, C., Perez, K., Praz, V., Lopez Garcia, G., Hinte, L. C., Meyenn, F., & Ocampo, A. (2024). Atac-clock: An aging clock based on chromatin accessibility. Geroscience, 46(2), 1789–1806. https://doi.org/10.1007/s11357-023-00986-0
Oh, H. S., Rutledge, J., Nachun, D., Palovics, R., Abiose, O., Moran-Losada, P., Channappa, D., Urey, D. Y., Kim, K., Sung, Y. J., Wang, L., Timsina, J., Western, D., Liu, M., Kohlfeld, P., Budde, J., Wilson, E. N., Guen, Y., Maurer, T. M., … Wyss-Coray, T. (2023). Organ aging signatures in the plasma proteome track health and disease. Nature, 624(7990), 164–172. https://doi.org/10.1038/s41586-023-06802-1
Oliva, M., Demanelis, K., Lu, Y., Chernoff, M., Jasmine, F., Ahsan, H., Kibriya, M. G., Chen, L. S., & Pierce, B. L. (2023). Dna methylation qtl mapping across diverse human tissues provides molecular links between genetic variation and complex traits. Nature Genetics, 55(1), 112–122. https://doi.org/10.1038/s41588-022-01248-z
Paszke, A., Gross, S., Massa, F., Lerer, A., Bradbury, J., Chanan, G., Killeen, T., Lin, Z., Gimelshein, N., & Antiga, L. (2019). Pytorch: An imperative style, high-performance deep learning library. In Advances in neural information processing systems (Vol. 32).
Peters, M. J., Joehanes, R., Pilling, L. C., Schurmann, C., Conneely, K. N., Powell, J., Reinmaa, E., Sutphin, G. L., Zhernakova, A., Schramm, K., Wilson, Y. A., Kobes, S., Tukiainen, T., Consortium, N. U., Ramos, Y. F., Goring, H. H., Fornage, M., Liu, Y., Gharib, S. A., Stranger, B. E., De Jager, P. L., Aviv, A., Levy, D., Murabito, J. M., Munson, P. J., Huan, T., Hofman, A., Uitterlinden, A. G., Rivadeneira, F., Rooij, J., Stolk, L., Broer, L., Verbiest, M. M., Jhamai, M., Arp, P., Metspalu, A., Tserel, L., Milani, L., Samani, N.J., Peterson, P., Kasela, S., Codd, V., Peters, A., Ward-Caviness, C.K., Herder, C., Waldenberger, M., Roden, M., Singmann, P., Zeilinger, S., Illig, T., Homuth, G., Grabe, H.J., Volzke, H., Steil, L., Kocher, T., Murray, A., Melzer, D., Yaghootkar, H., Bandinelli, S., Moses, E. K., Kent, J. W., Curran, J. E., Johnson, M.P., Williams-Blangero, S., Westra, H. J., McRae, A. F., Smith, J. A., Kardia, S. L., Hovatta, I., Perola, M., Ripatti, S., Salomaa, V., Henders, A. K., Martin, N. G., Smith, A. K., Mehta, D., Binder, E. B., Nylocks, K. M., Kennedy, E. M., Klengel, T., Ding, J., Suchy-Dicey, A. M., Enquobahrie, D. A., Brody, J., Rotter, J. I., Chen, Y. D., Houwing-Duistermaat, J., Kloppenburg, M., Slagboom, P. E., Helmer, Q., Hollander, W., Bean, S., Raj, T., Bakhshi, N., Wang, Q.P., Oyston, L. J., Psaty, B. M., Tracy, R. P., Montgomery, G. W., Turner, S. T., et al. (2015). The transcriptional landscape of age in human peripheral blood. Nature Communications, 6, 8570. https://doi.org/10.1038/ncomms9570
Ransohoff, J. D., Wei, Y., & Khavari, P. A. (2018). The functions and unique features of long intergenic non-coding rna. Nature Reviews Molecular Cell Biology, 19(3), 143–157. https://doi.org/10.1038/nrm.2017.104
Rattan, S. I. S. (2018). Biogerontology: Research status, challenges and opportunities. Acta Biomedica, 89(2), 291–301. https://doi.org/10.23750/abm.v89i2.7403
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., & Smyth, G. K. (2015). limma powers differential expression analyses for rna-sequencing and microarray studies. Nucleic Acids Research, 43(7), 47–47. https://doi.org/10.1093/nar/gkv007
Rivero-Segura, N. A., Bello-Chavolla, O. Y., Barrera-Vazquez, O. S., Gutierrez-Robledo, L. M., & Gomez-Verjan, J. C. (2020). Promising biomarkers of human aging: In search of a multi-omics panel to understand the aging process from a multidimensional perspective. Ageing Research Reviews, 64, 101164. https://doi.org/10.1016/j.arr.2020.101164
Robinson, O., Chadeau Hyam, M., Karaman, I., Climaco Pinto, R., Ala-Korpela, M., Handakas, E., Fiorito, G., Gao, H., Heard, A., Jarvelin, M. R., Lewis, M., Pazoki, R., Polidoro, S., Tzoulaki, I., Wielscher, M., Elliott, P., & Vineis, P. (2020). Determinants of accelerated metabolomic and epigenetic aging in a UK cohort. Aging Cell, 19(6), 13149. https://doi.org/10.1111/acel.13149
Rönn, T., Volkov, P., Gillberg, L., Kokosar, M., Perfilyev, A., Jacobsen, A. L., Jørgensen, S. W., Brøns, C., Jansson, P.-A., Eriksson, K.-F., Pedersen, O., Hansen, T., Groop, L., Stener-Victorin, E., Vaag, A., Nilsson, E., & Ling, C. (2015). Impact of age, bmi and hba1c levels on the genome-wide dna methylation and mrna expression patterns in human adipose tissue and identification of epigenetic biomarkers in blood. Human Molecular Genetics, 24(13), 3792–3813. https://doi.org/10.1093/hmg/ddv124
Sayed, N., Huang, Y., Nguyen, K., Krejciova-Rajaniemi, Z., Grawe, A. P., Gao, T., Tibshirani, R., Hastie, T., Alpert, A., Cui, L., Kuznetsova, T., Rosenberg-Hasson, Y., Ostan, R., Monti, D., Lehallier, B., Shen-Orr, S. S., Maecker, H. T., Dekker, C. L., Wyss-Coray, T., … Furman, D. (2021). An inflammatory aging clock (iage) based on deep learning tracks multimorbidity, immunosenescence, frailty and cardiovascular aging. Nature Aging, 1(7), 598–615. https://doi.org/10.1038/s43587-021-00082-y
Sengupta, S., & Brown, D. E.: Automatic report generation for histopathology images using pre-trained vision transformers. arXiv:2311.06176 (2023)
Shokhirev, M. N., & Johnson, A. A. (2021). Modeling the human aging transcriptome across tissues, health status, and sex. Aging Cell, 20(1), 13280. https://doi.org/10.1111/acel.13280
Slieker, R. C., Relton, C. L., Gaunt, T. R., Slagboom, P. E., & Heijmans, B. T. (2018). Age-related dna methylation changes are tissue-specific with elovl2 promoter methylation as exception. Epigenetics Chromatin, 11(1), 25. https://doi.org/10.1186/s13072-018-0191-3
Smith, Z. D., & Meissner, A. (2013). Dna methylation: Roles in mammalian development. Nature Reviews Genetics, 14(3), 204–220. https://doi.org/10.1038/nrg3354
Solovev, I., Shaposhnikov, M., & Moskalev, A. (2020). Multi-omics approaches to human biological age estimation. Mechanisms of Ageing and Development, 185, 111192. https://doi.org/10.1016/j.mad.2019.111192
Takahashi, S., Asada, K., Takasawa, K., Shimoyama, R., Sakai, A., Bolatkan, A., Shinkai, N., Kobayashi, K., Komatsu, M., Kaneko, S., Sese, J., & Hamamoto, R. (2020). Predicting deep learning based multi-omics parallel integration survival subtypes in lung cancer using reverse phase protein array data. Biomolecules. https://doi.org/10.3390/biom10101460
Tanaka, T., Biancotto, A., Moaddel, R., Moore, A. Z., Gonzalez-Freire, M., Aon, M. A., Candia, J., Zhang, P., Cheung, F., Fantoni, G., Consortium, C.H.I., Semba, R.D., & Ferrucci, L. (2018). Plasma proteomic signature of age in healthy humans. Aging Cell, 17(5), 12799. https://doi.org/10.1111/acel.12799
Urban, A., Sidorenko, D., Zagirova, D., Kozlova, E., Kalashnikov, A., Pushkov, S., Naumov, V., Sarkisova, V., Leung, G. H. D., Leung, H. W., Pun, F. W., Ozerov, I. V., Aliper, A., Ren, F., & Zhavoronkov, A. (2023). Precious1gpt: multimodal transformer-based transfer learning for aging clock development and feature importance analysis for aging and age-related disease target discovery. Aging (Albany NY), 15(11), 4649–4666. https://doi.org/10.18632/aging.204788
Viñuela, A., Brown, A. A., Buil, A., Tsai, P. C., Davies, M. N., Bell, J. T., Dermitzakis, E. T., Spector, T. D., & Small, K. S. (2018). Age-dependent changes in mean and variance of gene expression across tissues in a twin cohort. Human Molecular Genetics, 27(4), 732–741. https://doi.org/10.1093/hmg/ddx424
Wang, X., Wang, L., & Xiang, W. (2023). Mechanisms of ovarian aging in women: A review. Journal of Ovarian Research, 16(1), 67. https://doi.org/10.1186/s13048-023-01151-z
Wezyk, M., Spólnicka, M., Pośpiech, E., Pepłońska, B., Zbieć-Piekarska, R., Ilkowski, J., Styczyńska, M., Barczak, A., Zboch, M., Filipek-Gliszczynska, A., Skrzypczak, M., Ginalski, K., Kabza, M., Makałowska, I., Barcikowska-Kotowicz, M., Branicki, W., Żekanowski, C. (2018). Hypermethylation of trim59 and klf14 influences cell death signaling in familial Alzheimer’s disease. Oxidative Medicine and Cellular Longevity,2018, 6918797. https://doi.org/10.1155/2018/6918797
Xia, X., Chen, X., Wu, G., Li, F., Wang, Y., Chen, Y., Chen, M., Wang, X., Chen, W., Xian, B., Chen, W., Cao, Y., Xu, C., Gong, W., Chen, G., Cai, D., Wei, W., Yan, Y., Liu, K., … Han, J. J. (2020). Three-dimensional facial-image analysis to predict heterogeneity of the human ageing rate and the impact of lifestyle. Nature Metabolism, 2(9), 946–957. https://doi.org/10.1038/s42255-020-00270-x
Yang, X., Lay, F., Han, H., & Jones, P. A. (2010). Targeting dna methylation for epigenetic therapy. Trends in Pharmacological Sciences, 31(11), 536–46. https://doi.org/10.1016/j.tips.2010.08.001
Yang, Y., Zha, K., Chen, Y., Wang, H., Katabi, D. (2021). Delving into deep imbalanced regression. In International conference on machine learning (pp. 11842–11851). PMLR.
Zhang, Q. (2023). An interpretable biological age. The Lancet Healthy Longevity, 4(12), 662–663. https://doi.org/10.1016/S2666-7568(23)00213-1
Funding
Open access funding provided by FCT|FCCN (b-on). RR was supported by the scholarship BD/07092/2021 from Fundação para a Ciência e a Tecnologia (FCT) and the European Social Fund. MM was supported by the scholarship SFRH/BD/145707/2019 from FCT and the European Social Fund. PGF acknowledges the Grant 2022.15770.CPCA.A1 from RNCA-FCT.
Author information
Authors and Affiliations
Contributions
RR, MM and PGF contributed to the study conception and design. RR, MM and AM conducted the analysis. All authors contributed to the writing of the manuscript, read and approved the final version. The authors thank the reviewers for their thoughtful critiques, which helped us to improve the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare no Conflict of interest.
Additional information
Editors: Ana Carolina Lorena, Albert Bifet, Rita P. Ribeiro.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
This manuscript contains supplementary tables (Supplementary Table 1–5), submitted as an excel file, as well as supplementary methods submitted as part of the annex. The information sheet was also submitted in PDF format. Below is the link to the electronic supplementary material.
Appendix A Supplementary Methods
Appendix A Supplementary Methods
1.1 A.1 Train-test split and cross-validation
The methylation data set was split into train-test sets on a per-tissue basis. Since the data is skewed towards older ages, we took steps to ensure representation of younger individuals in the test set. We divided the dataset into younger (< 45 years) and older (> 45 years) individuals. For younger individuals, we performed a 50–50 stratified split (5 year intervals) between train-test sets. We then split the older samples, also using stratified splits, to total an 80–20 overall split between train-test. The test samples contained only individuals common between all 4 data modalities. For the remaining data modalities, samples not included in the common test set were used for model training.
1.2 A.2 Inference using molecular clocks
We applied the state-of-the-art epigenetic models to the train-test methylation sets separately. For the Horvath and Hannum clock, we downloaded the coefficients of each probe from the respective publications (Horvath, 2013; Hannum et al., 2013). These coefficients were then used to predict individual age through linear models. Since the Hannum clock was originally trained in whole blood samples, we recalibrated the model for other tissues, similarly to the author’s strategy. To that end, we first predicted the age in the train set, \(\hat{y}\), for each tissue using the Hannum clock. We then fit a simple linear regression model, \(y = \hat{y} \times b_1 + b_0\), where \(y\) represents the true age, \(b_1\) the regression coefficient on the predicted age in the Hannum clock and \(b_0\) the intersect. Then, we applied this regression to the predictions from the Hannum clock on the test set to obtain the calibrated predictions. Finally, for AltumAge, we first normalized the samples as delineated by the authors (Lima Camillo et al., 2022) and applied the model downloaded from the companion repository (https://github.com/rsinghlab/AltumAge).
Across all three implemented models, we found missing features in our dataset. Before model prediction, each set of missing features was imputed by taking the mean of \(\beta \)-values of a subset of features in each sample. For the models developed in Horvath (2013) and Lima Camillo et al. (2022), we considered the Illumina Infinium HumanMethylation27, while for the model developed in Hannum et al. (2013) we considered the Illumina Infinium HumanMethylation450. This choice was driven by the data used to train each model in the original publication. Data imputation was performed before the train-test split. It should be noted that missing value imputation might affect final prediction accuracy.
1.3 A.3 DNA methylation data processing
DNA methylation is measured in \(\beta \), with values ranging from 0 to 1 (Eq. A1):
where \(M\) and \(U\) denote the methylated and unmethylated signal intensities measured by the Illumina array. The offset \(a \ge 0\) is usually set equal to 100 and is added to \(M + U\) to stabilize beta values when both \(M\) and \(U\) are small. We converted these \(\beta \)-values to M-values, as the former are less prone to heteroscedasticity (Du et al., 2010). For this conversion, we followed the implementation of the R package ENmix presented in Eq. A2
1.4 A.4 Gene expression data pre-filtering
GTEx transcript read counts were available as TPM (transcripts per million), which normalizes for sample-wise sequencing depth and gene length. Equation A3 represents this transformation
where \(r_i\) represents the number of reads mapped to a gene and \(t_{li}\) represents transcript length. For each tissue, we filtered for protein-coding genes and long intergenic noncoding RNAs (lincRNAs). Research has shown that lincRNAs regulate key biological processes often involved in various diseases and are highly tissue-specific (Ransohoff et al., 2018). Chosen genes underwent additional filtering to select those with a minimum TPM of 1 in 20% of samples, ensuring the presence of significant biological signals. Only genes passing this filtering process were considered for feature selection.
1.5 A.5 Feature selection for molecular datasets
Feature selection was considered primarily due to the very large number of input features compared with the number of samples (i.e., \( p \gg n \)). We performed feature selection in the methylation dataset by identifying methylation features changing with age (differential methylation loci; DML) in each tissue, using the limma package (Ritchie et al., 2015). We corrected this analysis for Hardy scale, ischemic time, sex (in lung) and BMI. We ran the analysis using M values (Du et al., 2010). For the gene expression, we performed a similar feature selection method and identified differential expressed genes (DEG). We corrected this analysis for Hardy scale, ischemic time, sex (in lung), BMI and RIN values. Distribution of relevant covariates are presented in supplementary table 5.
We performed the analysis in a tissue-specific manner in both gene expression and epigenetic data modality and corrected for multiple testing using false discovery rate (FDR) through the Benjamini–Hochberg method. Probes and genes were considered DML and DEG at an adjusted p value below 0.05.
Elastic net combines both L1 and L2 penalization methods to shrink coefficients to 0, resulting in a sparse coefficient matrix of uncorrelated predictors. As such, it can also be considered as a feature selection method. We leveraged this property of ENs as an additional feature selection method during training of non-linear models (GBT and NN). To decrease model runtime, we precomputed folds for each tissue and performed feature selection using only the samples in the training folds. Furthermore, we performed feature selection using the whole training set for the final model training.
1.6 A.6 Addressing age data imbalance
In our previous work (Moraes et al., 2023), we identified the underrepresentation of younger ages in our training dataset as a significant factor that led to a reduced accuracy in our models, particularly for ages below 45 years. In an attempt to mitigate this problem, we implemented a technique known as Label Distribution Smoothing (LDS) (Yang et al., 2021), which employs a symmetric 1-dimensional kernel, such as Gaussian, Laplacian or triangular, to convolve with the empirical label distribution. This convolution produces an effective label density distribution that better represents the continuity of age labels. With this refined distribution, we can then adopt strategies typically used in classification tasks, such as calculating weights based on the inverse frequency of labels, which can be incorporated into the loss function during the training phase.
1.7 A.7 DNA methylation and gene expression model training
For both DNA methylation and gene expression model training we used EN and GBT (as implemented in lightGBM), following the sklearn and lightgbm python packages (Ke et al., 2017) respectively. Before model training we applied feature selection as outlined above, followed by quantile normalization and LDS (Yang et al., 2021). We optimized the pipeline following a fivefold age stratified cross-validation scheme using the optuna framework (Akiba et al., 2019). The best model of each type was selected based on the average MAE across a fivefold cross-validation schema in the train data. Optimized parameters included model hyperparameters (l1 ratio and alpha in EN; number of estimators, learning rate, L1 and L2 regularization, number of leaves, max depth, subsample, feature fraction and minimum child samples in GBT) and the use of LDS in the pipeline and respective hyperparameters (kernel, reweight, ks and sigma). Furthermore, for GBT models, we left the selection of feature selection methods to optuna (DML or EN). We identified the best combination of these parameters by using MAE as the selection criterion. We then trained the final model using the entire training dataset with the optimized parameters in order to assess the performance on the common test set.
1.8 A.8 Histology Feature Extraction Pipeline
Whole Slide Images (WSI) are gigapixel images that can be as large as \( 150,000 \times 150,000 \) pixels. Because of their high resolution and fixed scale, these images naturally present a hierarchical structure when considered from different levels of magnification, going from individual cells to complex tissue structures. Due to this large number of pixels, WSIs cannot be stored directly as tensors into the GPU’s memory, forcing us to break them down into small size patches in order to feed our neural network models, a process that leads us to contextual information loss across magnification levels.
To get around this problem, Chen et al. (2023) proposed a Visual Transformer (ViT) based architecture called Hierarchical Pyramid Image Transformer (HIPT) to extract meaningful features from three main WSI resolutions: \( [16 \times 16] \) (cellular features), \( [256 \times 256] \) (cellular organization), and \( [4096 \times 4096] \) (tissue phenotype).
In our previous work (Moraes et al., 2023) utilizing WSIs for chronological age detection, we hypothesized that a contributing factor to the reduced accuracy of the histological models was the loss of contextual information caused by the selection of \( [256 \times 256] \) pixel patches, which focused solely on cellular-level features, potentially overlooking broader tissue structures and contextual cues that might be useful for an accurate age determination. Therefore, in this study, we employed the HIPT architecture as our base model within a transfer learning framework, aiming to retain meaningful information from different magnification levels.
Using the same notation as in (Chen et al., 2023), \( x_L \) should be understood as an image of size \( [L \times L] \), and \( \{ x_l(i) \}_{i=1} \in \mathbb {R}^{M \times d} \) as a set of \( M \) visual tokens extracted from non-overlapping patches of size \( [l \times l] \) within \( x_L \), each one having a latent representation of \( d \) dimensions. Additionally, we should interpret \( \text {ViT}_L -l \) as a Vision Transformer that takes as input an image \( x_L \), processes each one of its \( x_l \) patches, \( l = \sqrt{L} \), and returns an embedding representation of dimension \( d \) for \( x_L \).
In order to utilize HIPT as a feature extractor for our downsampling task, we divided each WSI into \( M \) patches \( x_{4096} \) of size \( [4096 \times 4096] \), ending up with a set of patches \( \{ x_{4096}^{k} \}_{k=1}^M \) per WSI. Then, for each \( k \)-patch \( x_{4096}^{k} \), we extracted \( [CLS] \) tokens from the two main ViTs that compose the HIPT architecture, \( \text {ViT}_{256}\text {-16} \) and \( \text {ViT}_{4096}\text {-256} \), ending up with two types of visual tokens per \( x_{4096}^{k} \), \( [CLS]_{256}^{(j)} \in \mathbb {R}^{256 \times 384} \) and \( [CLS]_{4096}^{(k)} \in \mathbb {R}^{1 \times 192} \) respectively, where \( j \) is the index of the \( j \)-patch \( x_{256}^j \) within \( x_{4096}^k \).
Thus, for each WSI we have a set of \( M \) visual tokens \( \{ [CLS]_{4096}^{(k)} \}_{k=1} \in \mathbb {R}^{M \times 192} \), and a set of \( N \) visual tokens \( \{ [CLS]_{256}^{(j)} \}_{j=1}\in \mathbb {R}^{N \times 384} \), where \( M=35 \) on average and \( N=M \times 256 \) since there are always \( 256 \times 16 \) patches inside any \( x_{4096} \) image.
Finally, in order to get a WSI level representation, we took the mean of all \( \{ [CLS]_{4096}^{(k)} \}_{k=1}^M \) over the \( M \) dimension, \( [CLS]_{4096}^{WSI} = \frac{1}{M} \sum _{k=1}^M [CLS]_{4096}^{(k)} \in \mathbb {R}^{1 \times 384} \), and the mean of all \( \{ [CLS]_{256}^{(j)} \}_{j=1}^N \) over the \( N=256 \times M \) dimension, \( [CLS]_{256}^{WSI} = \frac{1}{M} \sum _{j=1}^M \left( \frac{1}{256} \sum _{i=1}^{256} [CLS]_{256}^{(i)} \right) \in \mathbb {R}^{1 \times 384} \), in both cases keeping the feature dimension constant. Additionally, we also concatenate \( [CLS]_{4096}^{WSI} \) with \( [CLS]_{256}^{WSI} \) in the feature axis to obtain a third representation, \( [CLS]_{4096,256}^{WSI} \in \mathbb {R}^{1 \times 576} \).
The code used in the beginning of this pipeline to divide the WSI into the \( x_{4096} \) patches were adapted from Sengupta and Brown (2023).
1.9 A.9 Histological images model training
Using the histological features extracted as explained in the previous section, we trained EN and NN models following the sklearn and pytorch (Paszke et al., 2019) python packages respectively. In the case of NN models we configured a Multilayer Perceptron (MLP) to behave as an encoder, forcing the dimensionality of its input data to be progressively reduced throughout the forward propagation process. Similar to the previous section, we used LDS (Yang et al., 2021) to mitigate the age skewness in the dataset. We optimized the pipeline following a fivefold age stratified cross-validation scheme using the optuna framework (Akiba et al., 2019). The best model of each type was selected based on the average MAE across a fivefold cross-validation schema in the train data. Optimized parameters included model hyperparameters (l1 ratio and alpha in EN; optimizer, learning rate, number of nodes, dropout rates, number of epochs and batch size in NN), the use or quantile normalization, the use of LDS in the and respective parameters (kernel, reweight, ks and sigma), and the selection of features set (\( [CLS]_{256}^{WSI} \), \( [CLS]_{4096}^{WSI} \) or \( [CLS]_{4096,256}^{WSI} \)). We identified the best combination of these parameters by using average MAE across the fivefolds as the selection criterion. The final model was trained using the best combination of parameters on the whole train set.
1.10 A.10 Telomere model training
For telomere model training we fitted multiple linear regression models where TL was combined with demographic and technical variables: BMI, sex (in lung) and ethnicity. We evaluated model performance using both a fivefold cross validation scheme and the test set.
1.11 A.11 Multi-modal model training
To train models using multi-modal data we used EN, GBT and NN following the sklearn, lightgbm (Ke et al., 2017) and pytorch (Paszke et al., 2019) python packages respectively.
Since not all samples are common between the four data modalities, we recomputed the folds for this experiment and performed feature selection for both DNA methylation and gene expression as previously described. For the selection of features to be used in the multi-modal models, we leveraged not only the samples in the training folds but also the modality-specific samples. Since these individuals are not present in the validation folds or in the common test set we are able to have a more robust feature selection process without data leakage. We also included these individuals when performing feature selection using the whole train set for training the final model.
For EN and GBT we combined the features of each data modality and optimized the pipeline using only the common samples in a fivefold cross-validation scheme as described in the previous sections. For NN models, we expanded the previous architecture used to deal with histological features by configuring multiple input layers, each one associated with a different modality and connected to its own custom MLP. For the gene expression and methylation features in each selection method we subsetted the common features in the fivefolds and ranked them based on effect size (differential analysis features) or based on coefficient size (EN embedded method features). We then selected the top features (128 for gene expression, 256 for methylation), which were used for model training. For each data modality (histology, gene expression and methylation data) we configured its associated MLP to behave as an encoder, followed by a fusion of the encoded features of each modality through a concatenation operation and, finally, a linear projection layer for age estimation. We optimized the parameters of this neural network using a fivefold cross-validation as previously described. The number of nodes, number of layers and dropout rates were optimized in each data modality. After optimization, we selected the top features (128 for gene expression, 256 for methylation) from the feature selection performed with the whole train set and trained the model using the optimized hyperparameters.
1.12 A.12 SHAP analysis
SHAP values represent each feature estimated average marginal contribution to a prediction with local additivity (Lundberg & Lee, 2017). Importantly, the sum of the approximated SHAP values amounts to the difference between the expected model prediction on a given set of background samples and the prediction for an observed sample. We compute selected models SHAP values using LinearExplainer function in the shap python package in order to understand the most important features.
We leveraged the additive nature of these values to estimated the contribution of each data type to model prediction. Let S be the matrix of SHAP values, where each row corresponds to an observation and each column to a feature from any of the three modalities. First, we computed the absolute value of each element in S, resulting in matrix \(A=abs(S)\). Then, for each column j in A, we calculated the mean of the absolute values to obtain the vector \(\varvec{{\mu }}\), where \(\mu _j\) represents the mean absolute SHAP value for feature j:
where m is the number patients we have in our test dataset. Next, to normalize \(\varvec{{\mu }}\), we first calculated the total sum \(\sigma \) of its elements \(\mu _j\),
ending up with a vector \(\varvec{{\nu }}\), where each of its element \(\nu _j\) represents the normalized importance of feature j as a percentage:
Finally, considering \(I_{\text {meth}}\), \(I_{\text {hist}}\), and \(I_{\text {gene}}\) respectivelly as the indices for Methylation, Histology, and Gene Expression within \(\varvec{{\nu }}\), we calculated the contribution of each modality by summing up the normalized values for each modality:
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ribeiro, R., Moraes, A., Moreno, M. et al. Integration of multi-modal datasets to estimate human aging. Mach Learn (2024). https://doi.org/10.1007/s10994-024-06588-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10994-024-06588-x