Abstract
Novelty detection is one of the classic problems in machine learning that has applications across several domains. This paper proposes a novel autoencoder based on Deep Gaussian Processes for novelty detection tasks. Learning the proposed model is made tractable and scalable through the use of random feature approximations and stochastic variational inference. The result is a flexible model that is easy to implement and train, and can be applied to general novelty detection tasks, including large-scale problems and data with mixed-type features. The experiments indicate that the proposed model achieves competitive results with state-of-the-art novelty detection methods.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Novelty detection is a fundamental task across numerous domains, with applications in data cleaning (Liu et al. 2004), fault detection and damage control (Dereszynski and Dietterich 2011; Worden et al. 2000), fraud detection related to credit cards (Hodge and Austin 2004) and network security (Pokrajac et al. 2007), along with several medical applications such as brain tumor (Prastawa et al. 2004) and breast cancer (Greensmith et al. 2006) detection. Novelty detection targets the recognition of anomalies in test data which differ significantly from the training set (Pimentel et al. 2014), so this problem is also known as “anomaly detection”. Challenges in performing novelty detection stem from the fact that labelled data identifying anomalies in the training set is usually scarce and expensive to obtain, and that very little is usually known about the distribution of such novelties. Meanwhile, the training set itself might be corrupted by outliers and this might impact the ability of novelty detection methods to accurately characterize the distribution of samples associated with a nominal behavior of the system under study. Furthermore, there are many applications, such as the ones that we study in this work, where the volume and heterogeneity of data might pose serious computational challenges to react to novelties in a timely manner and to develop flexible novelty detection algorithms. As an example, the Airline IT company Amadeus provides booking platforms handling millions of transactions per second, resulting in more than 3 million bookings per day and Petabytes of stored data. This company manages almost half of the flight bookings worldwide and is targeted by fraud attempts leading to revenue losses and indemnifications. Detecting novelties in such large volumes of data is a daunting task for a human operator; thus, an automated and scalable approach is truly desirable.
Because of the difficulty in obtaining labelled data and since the scarcity of anomalies is challenging for supervised methods (Japkowicz and Stephen 2002), novelty detection is normally approached as an unsupervised machine learning problem (Pimentel et al. 2014). The considerations above suggest some desirable scalability and generalization properties that novelty detection algorithms should have.
We have recently witnessed the rise of deep learning techniques as the preferred choice for supervised learning problems, due to their large representational power and the possibility to train these models at scale (LeCun et al. 2015); examples of deep learning techniques achieving state-of-the-art performance on a wide variety of tasks include computer vision (Krizhevsky et al. 2012), speech recognition (Hinton et al. 2012), and natural language processing (Collobert and Weston 2008). A natural question is whether such impressive results can extend beyond supervised learning to unsupervised learning and further to novelty detection. Deep learning techniques for unsupervised learning are currently actively researched on Kingma and Welling (2014) and Goodfellow et al. (2014), but it is still unclear whether these can compete with state-of-the-art novelty detection methods. We are not aware of recent surveys on neural networks for novelty detection, and the latest one we could find is almost 15 years old (Markou and Singh 2003) and misses the recent developments in this domain.
Key challenges with the use of deep learning methods in general learning tasks are (1) the necessity to specify a suitable architecture for the problem at hand and (2) the necessity to control their generalization. While various forms of regularization have been proposed to mitigate the overfitting problem and improve generalization, e.g., through the use of dropout (Srivastava et al. 2014; Gal and Ghahramani 2016), there are still open questions on how to devise principled ways of applying deep learning methods to general learning tasks. Deep Gaussian Processes (dgps) are ideal candidate to simultaneously tackle issues (1) and (2) above. dgps are deep nonparametric probabilistic models implementing a composition of probabilistic processes that implicitly allows for the use of an infinite number of neurons at each layer (Damianou and Lawrence 2013; Duvenaud et al. 2014). Also, their probabilistic nature induces a form of regularization that prevents overfitting, and allows for a principled way of carrying out model selection (Neal 1996). While dgps are particularly appealing to tackle general deep learning problems, their training is computationally intractable. Recently, there have been contributions in the direction of making the training of these models tractable (Bui et al. 2016; Cutajar et al. 2017; Bradshaw et al. 2017), and these are currently in the position to compete with Deep Neural Networks (dnns) in terms of scalability, accuracy, while providing superior quantification of uncertainty (Gal and Ghahramani 2016; Cutajar et al. 2017; Gal et al. 2017).
In this paper, we introduce an unsupervised model for novelty detection based on dgps in autoencoder configuration. We train the proposed dgp autoencoder (dgp-ae) by approximating the dgp layers using random feature expansions, and by performing stochastic variational inference on the resulting approximate model. The key features of the proposed approach are as follows: (1) dgp-aes are unsupervised probabilistic models that can model highly complex data distribution and offer a scoring method for novelty detection; (2) dgp-aes can model any type of data including cases with mixed-type features, such as continuous, discrete, and count data; (3) dgp-aes training does not require any expensive and potentially numerically troublesome matrix factorizations, but only tensor products; (4) dgp-aes can be trained using mini-batch learning, and could therefore exploit distributed and GPU computing; (5) dgp-aes training using stochastic variational inference can be easily implemented taking advantage of automatic differentiation tools, making for a very practical and scalable methods for novelty detection. Even though we leave this for future work, it is worth mentioning that dgp-aes can easily include the use of special representations based, e.g., on convolutional filters for applications involving images, and allow for end-to-end training of the model and the filters.
We compare dgp-aes with a number of competitors that have been proposed in the literature of deep learning to tackle large-scale unsupervised learning problems, such as Variational Autoencoders (vae) (Kingma and Welling 2014), Variational Auto-Encoded Deep Gaussian Process (vae-dgp) (Dai et al. 2016) and Neural Autoregressive Distribution Estimator (nade) (Uria et al. 2016). Through a series of experiments, where we also compare against state-of-the-art novelty detection methods such as Isolation Forest (Liu et al. 2008) and Robust Kernel Density Estimation (Kim and Scott 2012), we demonstrate that dgp-aes offer flexible modeling capabilities with a practical learning algorithm, while achieving state-of-the-art performance.
The paper is organized as follows: Sect. 2 introduces the problem of novelty detection and reviews the related work on the state-of-the-art. Section 3 presents the proposed dgp-ae for novelty detection, while Sects. 4 and 5 report the experiments and conclusions.
2 Novelty detection
Consider an unsupervised learning problem where we are given a set of input vectors \(X = [\mathbf {x}_1, \ldots , \mathbf {x}_n]^{\top }\). Novelty detection is the task of classifying new test points \(\mathbf {x}_*\), based on the criterion that they significantly differ from the input vectors X, that is the data available at training time. Such data is assumed to be generated by a different generative process and are called anomalies. Novelty detection is thus a one-class classification problem, which aims at constructing a model describing the distribution of nominal samples in a dataset. Unsupervised learning methods allow for the prediction on test data \(\mathbf {x}_*\); given a model with parameters \({\varvec{\theta }}\), define predictions as \(h(\mathbf {x}_* | X, {\varvec{\theta }})\). Assuming \(h(\mathbf {x}_* | X, {\varvec{\theta }})\) to be continuous, it is possible to interpret it as a means of scoring test points as novelties. The resulting scores allow for a ranking of test points \(\mathbf {x}_*\) highlighting the patterns which differ the most from the training data X. In particular, it is possible to define a threshold \(\alpha \) and flag a test point \(\mathbf {x}_*\) as a novelty when \(h(\mathbf {x}_* | X, {\varvec{\theta }}) > \alpha \).
After thresholding, it is possible to assess the quality of a novelty detection algorithm using scores proposed in the literature for binary classification. Based on a labelled testing dataset, where novelties and nominal cases are defined as positive and negative samples, respectively, we can compute the precision and recall metrics given in Eq. 1. True positives (TP) are examples correctly labelled as positives, false positives (FP) refer to negative samples incorrectly labelled as positives, while false negatives (FN) are positive samples incorrectly labelled as negatives.
In the remainder of this paper we are going to assess results of novelty detection methods by varying \(\alpha \) over the range of values taken by \(h(\mathbf {x}_* | X, {\varvec{\theta }})\) over a set of test points. When we vary \(\alpha \), we obtain a set of precision and recall measurements resulting in a curve. We can then compute the area under the precision–recall curve called the mean average precision (map), which is a relevant metric to compare the performance of novelty detection methods (Davis and Goadrich 2006). In practical terms, \(\alpha \) is chosen to strike an appropriate balance between accuracy in identifying novelties and a low level of false positives.
Novelty detection has been thoroughly investigated by theoretical studies (Pimentel et al. 2014; Hodge and Austin 2004). The evaluation of state-of-the-art methods was also reported in experimental papers (Emmott et al. 2016), including experiments on the methods scalability (Domingues et al. 2018) and resistance to the curse of dimensionality (Zimek et al. 2012). In one of the most recent surveys on novelty detection (Pimentel et al. 2014), methods have been classified into the following categories. (1) Probabilistic approaches estimate the probability density function of X defined by the model parameters \({\varvec{\theta }}\). Novelties are scored by the likelihood function \(P(\mathbf {x}_* | {\varvec{\theta }})\), which computes the probability for a test point to be generated by the trained distribution. These approaches are generative, and provide a simple understanding of the underlying data through parameterized distributions. (2) Distance-based methods compute the pairwise distance between samples using various similarity metrics. Patterns with a small number of neighbors within a specified radius, or distant from the center of dense clusters of points, receive a high novelty score. (3) Domain-based methods learn the domain of the nominal class as a decision boundary. The label assigned to test points is then based on their location with respect to the boundary. (4) Information theoretic approaches measure the increase of entropy induced by including a test point in the nominal class. As an alternative, (5) isolation methods target the isolation of outliers from the remaining samples. As such, these techniques focus on isolating anomalies instead of profiling nominal patterns. (6) Most suitable unsupervised neural networks for novelty detection are autoencoders, i.e., networks learning a compressed representation of the training data by minimizing the error between the input data and the reconstructed output. Test points showing a high reconstruction error are labelled as novelties. Our model belongs to this last category, and extends it by proposing a nonparametric and probabilistic approach to alleviate issues related to the choice of a suitable architecture while accounting for the uncertainty in the autoencoder mappings; crucially, we show that this can be achieved while learning the model at scale.
3 Deep Gaussian Process autoencoders for novelty detection
In this section, we introduce the proposed dgp-ae model and describe the approximation that we use to make inference tractable and scalable. Each iteration of the algorithm is linear in the dimensionality of the input, batch size, dimensionality of the latent representation and number of Monte Carlo samples used in the approximation of the objective function, which highlights the tractability of the model. We also discuss the inference scheme based on stochastic variational inference, and show how predictions can be made. Finally, we present ways in which we can make the proposed dgp-ae model handle various types of data, e.g., mixing continuous and categorical features. We refer the reader to Cutajar et al. (2017) for a detailed derivation of the random feature approximation of dgps and variational inference of the resulting model. In this work, we extend this dgp formulation to autoencoders.
3.1 Deep Gaussian Process autoencoders
An autoencoder is a model combining an encoder and a decoder. The encoder part takes each input \(\mathbf {x}\) and maps it into a set of latent variables \(\mathbf {z}\), whereas the decoder part maps latent variables \(\mathbf {z}\) into the inputs \(\mathbf {x}\). Because of their structure, autoencoders are able to jointly learn latent representations for a given dataset and a model to produce \(\mathbf {x}\) given latent variables \(\mathbf {z}\). Typically this is achieved by minimizing a reconstruction error.
Autoencoders are not generative models, and variational autoencoders have recently been proposed to enable this feature (Dai et al. 2016; Kingma and Welling 2014). In the context of novelty detection, the possibility to learn a generative model might be desirable but not essential, so in this work we focus in particular on autoencoders. Having said that, we believe that extending variational autoencoders using the proposed framework is possible, as well as empowering the current model to enable generative modeling; we leave these avenues of research for future work. In this work, we propose to construct the encoder and the decoder functions of autoencoders using dgps. As a result, we aim at jointly learning a probabilistic nonlinear projection based on dgps (the encoder) and a dgp-based latent variable model (the decoder).
The building block of dgps are gps, which are priors over functions; formally, a gp is a set of random variables characterized by the property that any subset of them is jointly Gaussian (Rasmussen and Williams 2006). The gp covariance function models the covariance between the random variables at different inputs, and it is possible to specify a parametric function for their mean.
Stacking multiple gps into a dgp means feeding the output of gps at each layer as the input of the gps at the next; this construction gives rise to a composition of stochastic processes. Assume that we compose \(N_{\mathrm {L}}\) possible functions modelled as multivariate gps, the resulting composition takes the form
Without loss of generality, we are going to assume that the gp s at each layer have zero mean, and that gp covariances at layer (l) are parameterized through a set of parameters \({\varvec{\theta }}^{(l)}\) shared across gps in the same layer.
Denote by \(F^{(i)}\) the collection of the multivariate functions \(\mathbf {f}^{(i)}\) evaluated at the inputs \(F^{(i-1)}\), and define \(F^{(0)} := X\). The encoder part of the proposed dgp-ae model maps the inputs X into a set of latent variables \(Z := F^{(j)}\) through a dgp, whereas the decoder is another dgp mapping Z into X. The dgp controlling the decoding part of the model, assumes a likelihood function that allows one to express the likelihood of the observed data X as \(p\left( X | F^{(N_{\mathrm {L}})}, {\varvec{\theta }}^{(N_{\mathrm {L}})}\right) \). The likelihood reflects the choice on the mappings between latent variables and the type of data being modelled, and it can include and mix various types and dimensionality; Sect. 3.5 discusses this in more detail.
By performing Bayesian inference on the proposed dgp-ae model we aim to integrate out latent variables at all layers, effectively integrating out the uncertainty in all the mappings in the encoder/decoder and the latent variables Z themselves. Learning and making predictions with dgp-aes, however, require being able to solve intractable integrals. To evaluate the marginal likelihood expressing the probability of observed data given model parameters, we need to solve the following
A similar intricate integral can be derived to express the predictive probability \(p(\mathbf {x}_* | X, {\varvec{\theta }})\). For any nonlinear covariance function, these integrals are intractable. In the next section, we show how random feature expansions of the gps at each layer expose an approximate model that can be conveniently learned using stochastic variational inference, as described in Cutajar et al. (2017).
3.2 Random feature expansions for dgp-aes
To start with, consider a shallow multivariate gp and denote by F the latent variables associated with the inputs. For a number of gp covariance functions, it is possible to obtain a low-rank approximation of the processes through the use of a finite set of basis functions, and transform the multivariate gp into a Bayesian linear model. For example, in the case of an rbf covariance function of the form
it is possible to employ standard Fourier analysis to show that \(k_{\mathrm {rbf}}\) can be expressed as an expectation under a distribution over spectral frequencies, that is:
After standard manipulation, it is possible to obtain an unbiased estimate of the integral above by mean of a Monte Carlo average:
where \(\mathbf {z}(\mathbf {x}| \varvec{\omega }) = [\cos (\mathbf {x}^{\top } \varvec{\omega }), \sin (\mathbf {x}^{\top } \varvec{\omega })]^{\top }\) and \(\tilde{\varvec{\omega }}_{r} \sim p(\varvec{\omega })\). It is possible to increase the flexibility of the rbf covariance above by scaling it by a marginal variance parameter \(\sigma ^2\) and by scaling the features individually with length-scale parameters \(\varLambda = \mathrm {diag}(l_1^2,\ldots ,l_{DF}^2)\); it is then possible to show that \(p(\varvec{\omega }) = \mathcal {N}\left( \varvec{\omega }| \mathbf {0}, \varLambda ^{-1} \right) \) using Bochner’s theorem. By stacking the samples from \(p(\varvec{\omega })\) by column into a matrix \(\varOmega \), we can define
where the functions \(\cos ()\) and \(\sin ()\) are applied element-wise. We can now derive a low-rank approximation of K as follows:
It is straightforward to verify that the individual columns of F in the original gp can be approximated by the Bayesian linear model \(F_{\cdot j} = \varPhi W_{\cdot j}\) with \(W_{\cdot j} \sim \mathcal {N}(\mathbf {0}, I)\), as the covariance of \(F_{\cdot j}\) is indeed \(\varPhi \varPhi ^{\top } \approx K\).
The decomposition of the gp covariance in Eq. 4 suggests an expansion with an infinite number of basis functions, thus leading to a well-known connection with single-layered neural networks with infinite neurons (Neal 1996); the random feature expansion that we perform using Monte Carlo induces a truncation of the infinite expansion. Based on the expansion defined above, we can now build a cascade of approximate gps, where the output of layer l becomes the input of layer \(l+1\). The layer \(\varPhi ^{(0)}\) first expands the input features in a high-dimensional space, followed by a linear transformation parameterized by a weight matrix \(W^{(0)}\) which results in the latent variables \(F^{(1)}\) in the second layer. Considering a dgp with rbf covariances obtained by stacking the hidden layers previously described, we obtain Eqs. 9 and 10 derived from Eq. 6. These transformations are parameterized by prior parameters \((\sigma ^2)^{(l)}\) which determine the marginal variance of the gps and \(\varLambda ^{(l)} = \mathrm {diag}\left( \left( l_1^2\right) ^{(l)},\ldots ,\left( l_{DF^{(l)}}^2\right) ^{(l)}\right) \) describing the length-scale parameters.
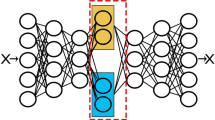
This leads to the proposed dgp-ae model’s topology given in Fig. 1. The resulting approximate dgp-ae model is effectively a Bayesian dnn where the priors for the spectral frequencies \(\varOmega ^{(l)}\) are controlled by covariance parameters \({\varvec{\theta }}^{(l)}\), and the priors for the weights \(W^{(l)}\) are standard normal.
Architecture of a 2-layer dgp autoencoder. Gaussian processes are approximated by hidden layers composed of two inner layers, the first layer \(\varPhi ^{(l)}\) performing random feature expansion followed by a linear transformation resulting in \(F^{(l)}\). Covariance parameters are \(\theta ^{(l)} = \left( (\sigma ^2)^{(l)}, \varLambda ^{(l)}\right) \), with prior over the weights \(p\left( \varOmega ^{(l)}_{\cdot j}\right) = N\left( 0, \big (\varLambda ^{(l)}\big )^{-1}\right) \) and \(p\left( W^{(l)}_{\cdot i}\right) = N(0, I)\). Z is the latent variables representation
In our framework, the choice of the covariance function induces different basis functions. For example, a possible approximation of the arc-cosine kernel (Cho and Saul 2009) yields Rectified Linear Units (relu) basis functions (Cutajar et al. 2017) resulting in faster computations compared to the approximation of the rbf covariance, given that derivatives of relu basis functions are cheap to evaluate.
3.3 Stochastic variational inference for dgp-aes
Let \({\varvec{\Theta }}\) be the collection of all covariance parameters \({\varvec{\theta }}^{(l)}\) at all layers; similarly, define \({\varvec{\Omega }}\) and \(\mathbf {W}\) to be the collection of the spectral frequencies \(\varOmega ^{(l)}\) and weight matrices \(W^{(l)}\) at all layers, respectively. We are going to apply stochastic variational inference techniques to infer \(\mathbf {W}\) and optimize all covariance parameters \({\varvec{\Theta }}\); we are going to consider the case where the spectral frequencies \({\varvec{\Omega }}\) are fixed, but these can also be learned (Cutajar et al. 2017). The marginal likelihood \(p(X | {\varvec{\Omega }}, {\varvec{\Theta }})\) can be bounded using standard variational inference techniques, following Kingma and Welling (2014) and Graves (2011), Defining \( \mathcal {L}= \log \left[ p(X | {\varvec{\Omega }}, {\varvec{\Theta }})\right] \), we obtain
Here the distribution \(q(\mathbf {W})\) denotes an approximation to the intractable posterior \(p(\mathbf {W}| X, {\varvec{\Omega }}, {\varvec{\Theta }})\), whereas the prior on \(\mathbf {W}\) is the product of standard normal priors resulting from the approximation of the gp s at each layer \( p(\mathbf {W}) = \prod _{l=0}^{N_{\mathrm {L}} - 1} p(W^{(l)}).\)
We are going to assume an approximate Gaussian distribution that factorizes across layers and weights
We are interested in finding an optimal approximate distribution \(q(\mathbf {W})\), so we are going to introduce the variational parameters \(m^{(l)}_{ij}, (s^2)^{(l)}_{ij}\) to be the mean and the variance of each of the approximating factors. Therefore, we are going to optimize the lower bound above with respect to all variational parameters and covariance parameters \({\varvec{\Theta }}\).
Because of the chosen Gaussian form of \(q(\mathbf {W})\) and given that the prior \(p(\mathbf {W})\) is also Gaussian, the DKL term in the lower bound to \(\mathcal {L}\) can be computed analytically. The remaining term in the lower bound, instead, needs to be estimated. Assuming a likelihood that factorizes across observations, it is possible to perform a doubly-stochastic approximation of the expectation in the lower bound so as to enable scalable stochastic gradient-based optimization. The doubly-stochastic approximation amounts to replacing the sum over n input points with a sum over a mini-batch of m points selected randomly from the entire dataset:
Then, each element of the sum can itself be estimated unbiasedly using Monte Carlo sampling and averaging, with \(\tilde{\mathbf {W}}_r \sim q(\mathbf {W})\):
Because of the unbiasedness property of the last expression, computing its derivative with respect to the variational parameters and \({\varvec{\Theta }}\) yields a so-called stochastic gradient that can be used for stochastic gradient-based optimization. The appeal of this optimization strategy is that it is characterized by theoretical guarantees to reach local optima of the objective function (Robbins and Monro 1951). Derivatives can be conveniently computed using automatic differentiation tools; we implemented our model in TensorFlow (Abadi et al. 2015) that has this feature built-in. In order to take derivatives with respect to the variational parameters we employ the so-called reparameterization trick (Kingma and Welling 2014)
to fix the randomness when updating the variational parameters, and \(\epsilon ^{(l)}_{rij}\) are resampled after each iteration of the optimization.
3.4 Predictions with dgp-aes
The predictive distribution for the proposed dgp-ae model requires solving the following integral
which is intractable due to fact that the posterior distribution over \(\mathbf {W}\) is unavailable. Stochastic variational inference yields an approximation \(q(\mathbf {W})\) to the posterior \(p(\mathbf {W}| X, {\varvec{\Omega }}, {\varvec{\Theta }})\), so we can use it to approximate the predictive distribution above:
where we carried out a Monte Carlo approximation by drawing \(N_{\mathrm {MC}}\) samples \(\tilde{\mathbf {W}}_r \sim q(\mathbf {W})\). The overall complexity of each iteration is thus \(\mathcal {O}\left( m D_F^{(l-1)} N_{RF}^{(l)} N_{MC}\right) \) to construct the random features at layer l and \(\mathcal {O}\left( mN_{RF}^{(l)}D_F^{(l)}N_{MC}\right) \) to compute the value of the latent functions at layer l, where m is the batch size and \(D_F^{(l)}\) is the dimensionality of \(F^{(l)}\). Hence, by carrying out updates using mini-batches, the complexity of each iteration is independent of the dataset size.
For a given test set \(X_*\) containing multiple test samples, it is possible to use the predictive distribution as a scoring function to identify novelties. In particular, we can rank the predictive probabilities \(p(\mathbf {x}_* | X, {\varvec{\Omega }}, {\varvec{\Theta }})\) for all test points to identify the ones that have the lowest probability under the given dgp-ae model. In practice, for numerical stability, our implementation uses log-sum operations to compute \(\log [p(\mathbf {x}_* | X, {\varvec{\Omega }}, {\varvec{\Theta }})]\), and we use this as the scoring function.
3.5 Likelihood functions
One of the key features of the proposed model is the possibility to model data containing a mix of types of features. In order to do this, all we need to do is to specify a suitable likelihood for the observations given the latent variables at the last layer, that is \(p(\mathbf {x}| \mathbf {f}^{(N_{\mathrm {L}})})\). Imagine that the vector \(\mathbf {x}\) contains continuous and categorical features that we model using Gaussian and multinomial likelihoods; extensions to other combinations of features and distributions is straightforward. Consider a single continuous feature of \(\mathbf {x}\), say \(x_{[G]}\); the likelihood function for this feature is:
For any given categorical feature, instead, assuming a one-hot encoding, say \(\mathbf {x}_{[C]}\), we can use a multinomial likelihood with probabilities given by the softmax transformation of the corresponding latent variables:
It is now possible to combine any number of these into the following likelihood function:
Any extra parameters in the likelihood function, such as the variances in the Gaussian likelihoods, can be included in the set of all model parameters \({\varvec{\Theta }}\) and learned jointly with the rest of parameters. For count data, it is possible to use the Binomial or Poisson likelihood, whereas for positive continuous variables we can use Exponential or Gamma. It is also possible to jointly model multiple continuous features and use a full covariance matrix for multivariate Gaussian likelihoods, multivariate Student-T, and the like. The nice feature of the proposed dgp-ae model is that the training procedure is the same regardless of the choice of the likelihood function, as long as the assumption of factorization across data points holds.
4 Experiments
We evaluate the performance of our model by monitoring the convergence of the mean log-likelihood (mll) and by measuring the area under the Precision–Recall curve, namely the mean average precision (map). These metrics are taken on real-world datasets described in Sect. 4.2. In addition, we compare our model against state-of-the-art neural networks suitable for outlier detection and highlighted in Sect. 4.1. To demonstrate the value of our proposal as a competitive novelty detection method, we include top performance novelty detection methods from other domains, namely Isolation Forest (Liu et al. 2008) and Robust Kernel Density Estimation (rkde) (Kim and Scott 2012), which are recommended for outlier detection in Emmott et al. (2016).
4.1 Selected methods
In order to retrieve a continuous score for the outliers and be able to compare the convergence of the likelihood for the selected models, our comparison focuses on probabilistic neural networks. Our dgp-ae is benchmarked against the Variational Autoencoder (vae) (Kingma and Welling 2014) and the Neural Autoregressive Distribution Estimator (nade) (Uria et al. 2016). We also include standard dnn autoencoders with sigmoid activation functions and dropout regularization to give a wider context to the reader. We initially intended to include Real nvp (Dinh et al. 2016) and Wasserstein gan (Arjovsky et al. 2017), but we found these networks and their implementations tightly tailored to images. The one-class classification with gps recently developed (Kemmler et al. 2013) is actually a supervised learning task where the authors regress on the labels and use heuristics to score novelties. Since this work is neither probabilistic nor a neural network, we did not include it. Parameter selection for the following methods was achieved by grid-search and maximized the average map over testing datasets labelled for novelty detection and described in Sect. 4.2. We append the depth of the networks as a suffix to the name, e.g., vae-2.
dgp -ae -g , dgp -ae -gs We train the proposed dgp-ae model for 100,000 iterations using 100 random features at each hidden layer. Due to the network topology, we use a number of multivariate gps equal to the number of input features when using a single-layer configuration, but use a multivariate gp of dimension 3 for the latent variables representation when using more than one layer. In the remainder of the paper, the term layer describes a hidden layer composed of two inner layers \(\varPhi ^{(i)}\) and \(F^{(i+1)}\). As observed in Duvenaud et al. (2014) and Neal (1996), deep architectures require to feed the input forward to the hidden layers in order to implement the modeling of meaningful functions. In the experiments involving more than 2 layers, we follow this advice by feed-forwarding the input to the encoding layers and feed-forward the latent variables to the decoding layers. The weights are optimized using a batch size of 200 and a learning rate of 0.01. The parameters \(q({\varvec{\Omega }})\) and \({\varvec{\Theta }}\) are fixed for 1000 and 7000 iterations respectively. \(N_{\mathrm {MC}}\) is set to 1 during the training, while we use \(N_{\mathrm {MC}} = 100\) at test time to score samples with higher accuracy. dgp-ae-g uses a Gaussian likelihood for continuous and one-hot encoded categorical variables. dgp-ae-gs is a modified dgp-ae-g where categorical features are modelled by a softmax likelihood as previously described. These networks use an rbf covariance function, except when the arc suffix is used, e.g., dgp-ae-g-1-arc.
vae-dgp-2Footnote 1 This network performs inducing points approximation to train a dgp model with variational inference. The network uses 2 hidden layers of dimensionality \(max(\frac{d}{2}, 5)\) and \(max(\frac{d}{3}, 4)\), and is trained for 1000 iterations over all training samples. All layers use a rbf kernel with 40 inducing points. The MLP in the recognition model has two layers of dimensionality 300 and 150.
vae-dgp,Footnote 2 vae-2 The variational autoencoder is a generative model which compresses the representation of the training data into a layer of latent variables, optimized with stochastic gradient descent. The sum of the reconstruction error and the latent loss, i.e., the negative of the Kullback–Leibler divergence between the approximate posterior over the latent variables and the prior, gives the loss term optimized during the training. The networks were trained for 4000 iterations using 50 hidden units and a batch size of 1000 samples. A learning rate of 0.001 was selected to optimize the weights. vae-1 is a shallow network using one layer for latent variables representation, while vae-2 uses a two-layer architecture with a first layer for encoding and a second one for decoding, each containing 100 hidden units. We use the reconstruction error to score novelties.
nade-2Footnote 3 This neural network is an autoencoder suitable for density estimation. The network uses mixtures of Gaussians to model p(x). The network yields an autoregressive model, which implies that the joint distribution is modelled such that the probability for a given feature depends on the previous features fed to the network, i.e., \(p(\varvec{x}) = p(x_{o_d}|\varvec{x}_{o_{<d}})\), where \(x_{o_d}\) is the feature of index d of \(\varvec{x}\). We train a deep and orderless nade for 5000 iterations using batches of 200 samples, a learning rate of 0.005 and a weight decay of 0.02. Training the network for more iterations increases the risk of the training failing due to runtime errors. The network has a 2 layer-topology with 100 hidden units and a relu activation function. The number of components for the mixture of Gaussians was set to 20, and we use Bernoulli distributions instead of Gaussians to model datasets exclusively composed of categorical data. 15% of the training data was used for validation to select the final weights.
ae-1, ae-5 These two neural networks are feedforward autoencoders using sigmoid activation functions in the hidden layers and a dropout rate of 0.5 to provide regularization. The first network is a single layer autoencoder with a number of hidden units equal to the number of features, while the second one has a 5-layer topology with 80% of the number of input features on the second and fourth layer, and 60% on the third layer. The networks are trained for 100,000 iterations with a batch size of 200 samples and a learning rate of 0.01. The reconstruction error is used to detect outliers.
Isolation forestFootnote 4 is a random forest algorithm performing recursive random splits over the feature domain until each sample is isolated from the rest of the dataset. As a result, outliers are separated after few splits and are located in nodes close to the root of the trees. The average path length required to reach the node containing the specified point is used for scoring. A contamination rate of 5% was used for this experiment.
rkdeFootnote 5 is a probabilistic method which assigns a kernel function to each training sample, then sums the local contribution of each function to give an estimate of the density. The experiment uses the cross-validation bandwidth (lkcv) as a smoothing parameter on the shape of the density, and the Huber loss function to provide a robust estimation of the maximum likelihood.
4.2 Datasets
Our evaluation is based on 11 datasets, including 7 datasets made publicly available by the UCI repository (Bache and Lichman 2013), while the 4 other datasets are proprietary datasets containing production data from the company Amadeus. This company provides online platforms to connect the travel industry and manages almost half of the flight bookings worldwide. Their business is targeted by fraud attempts reported as outliers in the corresponding datasets. The proprietary datasets are given thereafter; pnr describes the history of changes applied to booking records, transactions depicts user sessions performed on a Web application and targets the detection of bots and malicious users, shared-access was extracted from a backend application dedicated to shared rights management between customers, e.g., seat map display or cruise distribution, and payment-sub reports the booking records along with the user behavior through the booking process, e.g., searches and actions performed. Table 1 shows the datasets characteristics.
4.3 Results
This section shows the outlier detection capabilities of the methods and monitors the mll to exhibit convergence. We also study the impact of depth and dimensionality on dgp-aes, and plot the latent representations learnt by the network.
4.3.1 Method comparison
Our experiment performs a fivefold Monte Carlo cross-validation, using 80% of the original dataset for the training and 20% for the testing. Training and testing datasets are normalized, and we use the characteristics of the training dataset to normalize the testing data. Both datasets contain the same proportion of anomalies. Since class distribution is by nature heavily imbalanced for novelty detection problems, we use the map as a performance metric instead of the average rocauc. Indeed, the precision metric strongly penalizes false positives, even if they only represent a small proportion of the negative class, while false positives have very little impact on the roc (Davis and Goadrich 2006). The detailed map are reported in Table 2. Bold results are similar to the best map achieved on the dataset with nonsignificant differences. We used a pairwise Friedman test (Garca et al. 2010) with a threshold of 0.05 to reject the null hypothesis. The experiments are performed on an Ubuntu 14.04 LTS powered by an Intel Xeon E5-4627 v4 CPU and 256 GB RAM. This amount of memory is not sufficient to train rkde on the airline dataset, resulting in missing data in Table 2.
Looking at the average performance, our dgps autoencoders achieve the best results for novelty detection. dgps performed well on all datasets, including high dimensional cases, and outperform the other methods on wine-quality, airline and pnr. By fitting a softmax likelihood instead of a Gaussian on one-hot encoded features, dgp-ae-gs-1 achieves better performance than dgp-ae-g-1 on 3 datasets containing categorical variables out of 4, e.g., mushroom-sub, german-sub and transactions, while showing similar results on the car dataset. This representation allows dgps to reach the best performance on half of the datasets and to outperform state-of-the-art algorithms for novelty detection, such as rkde and IForest. Despite the low-dimensional representation of the latent variables, dgp-ae-g-2 achieves performance comparable to dgp-ae-g-1, which suggests good dimensionality reduction abilities. The use of a softmax likelihood in dgp-ae-gs-2 resulted in better novelty detection capabilities than dgp-ae-g-2 on the 4 datasets containing categorical features. vae-dgp-2 achieves good results but is outperformed on most small datasets.
vae-1 also shows good outlier detection capabilities and handles binary features better than vae-2. However, the multilayer architecture outperforms its shallow counterpart on large datasets containing more than 10,000 samples. Both algorithms perform better than nade-2 which fails on high dimensional datasets such as mushroom-sub, pnr or transactions. We performed additional tests with an increased number of units for nade-2 to cope for the large dimensionality, but we obtained similar results.
While ae-1 shows unexpected detection capabilities for a very simple model, ae-5 reaches the lowest performance. Compressing the data to a feature space 40% smaller than the input space along with dropout layers may cause loss of information resulting in an inaccurate model.
4.3.2 Convergence monitoring
To assess the accuracy and the scalability of the selected neural networks, we measure the map and mean log-likelihood (mll) on test data during the training phase to monitor their convergence. The evolution of the two metrics for the dnns is reported in Fig. 2.
While the likelihood is the objective function of most networks, the monitoring of this metric reveals occasional decreases of the mll for all methods during the training process. If minor increases are part of the gradient optimization, the others indicate convergence issues for complex datasets. This is observed for vae-dgp-2 and vae-1 on mammography, or dgp-ae-g-1-arc and vae-1 on mushroom-sub.
Our dgps show the best likelihood on most datasets, in particular when using the arc kernel, with the exception of pnr and mushroom-sub where the rbf kernel is much more efficient. These results demonstrate the efficiency of regularization for dgps and their excellent ability to generalize while fitting complex models.
On the opposite, nade-2 barely reaches the likelihood of ae-1 and ae-5 at convergence. In addition, the network requires an extensive tuning of its parameters and has a computationally expensive prediction step. We tweaked the parameters to increase the model complexity, e.g., number of components and units, but it did not improve the optimized likelihood.
vae-dgp-2 does not reach a competitive likelihood, even with deeper architectures, and shows a computationally expensive prediction step.
Looking at the overall results of these networks, we observe that the model, depicted here by the likelihood, is refined during the entire training process, while the average precision quickly stabilizes. This behavior implies that the ordering of data points according to their outlier score converges much faster, even though small changes can still occur.
Additional convergence experiments have been performed in dgps and are reported in Fig. 3. The left part of the figure shows the ability of dgp-ae-g to generalize while increasing the number of layers. On the right, we compare the dimensionality reduction capabilities of dgp-ae-g-2 while increasing the number of gps on the latent variables layer.
Evolution of the map and mll over time on testing data based on a threefold cross-validation process. The left plot reports the metrics for dgp-ae-g with an increasing number of layers. For networks with more than 2 layers, we feed forward the input to the encoding layers, and feed forward the latent variables to the decoding layers. We use 3 gps per layer and a length-scale of 1. The right plot shows the impact of an increasing number of gp nodes on a dgp-ae-g-2
The left part of the plot reports the convergence of dgp-ae-g for configurations ranging from one to ten layers. The plot highlights the correlation between a higher test likelihood and a higher average precision. Single-layer models show a good convergence of the mll on most datasets, though are outperformed by deeper models, especially 4-layer networks, on magic-gamma-sub, payment-sub and airline. Deep architectures result in models of higher capacity at the cost of needing larger datasets to be able to model complex representations, with a resulting slower convergence behavior. Using moderately deep networks can thus show better results on datasets where a single layer is not sufficient to capture the complexity of the data. Interestingly, the bound on the model evidence makes it possible to carry out model selection to decide on the best architecture for the model at hand (Cutajar et al. 2017).
In the right panel of Fig. 3, we increase the dimensionality of the latent representation fixing the architecture to a dgp-ae-g-2. Both the test likelihood and the average precision show that a univariate gp is not sufficient to model accurately the input data. The limitations of this configuration is observed on mammography, payment-sub and airline where more complex representations achieve better performance. Increasing the number of gps results in a higher number of weights for the model, thus in a slower convergence. While configurations using 5 GPs already perform a significant dimensionality reduction, they achieve good performance and are suitable for efficient novelty detection.
4.3.3 Latent representation
In this section we illustrate the capabilities of the proposed dgp-ae model to construct meaningful latent probabilistic representations of the data. We select a two-layer dgp-ae architecture with a two-dimensional latent representation \(Z := F^{(1)}\). Since the mapping of the dgp-ae model is probabilistic, each input point is mapped into a cloud of latent variables. In order to obtain a generative model, we could then train a density estimation algorithm on the latent variables to construct a density \(q(\mathbf {z})\) used together with the probabilistic decoder part of the dgp-ae to generate new observations.
In Fig. 4, we draw 300 Monte Carlo samples from the approximate posterior over the weights \(\mathbf {W}\) to construct a latent representation of the old faithful dataset. We use a gmm with two components to cluster the input data, and color the latent representation based on the resulting labels. The point highlighted on the left panel of the plot by a cross is mapped into the green points on the right.
We now extend our experiment to labelled datasets of higher dimensionality, using the given labels for the sole purpose of assigning a color to the points in the latent space. Figure 5 shows the two-dimensional representation of four datasets, breast cancer (569 samples, 30 features), iris (150 \(\times \) 4), wine (178 \(\times \) 13) and digits (1797 \(\times \) 64). For comparison, we also report the results of two manifold learning algorithms, namely t-sne (Maaten and Hinton 2008) and Probabilistic pca (Tipping and Bishop 1999). The plot shows that our algorithm yields meaningful low-dimensional representations, comparable with state-of-the-art dimensionality reduction methods.
5 Conclusions
In this paper, we introduced a novel deep probabilistic model for novelty detection. The proposed dgp-ae model is an autoencoder where the encoding and the decoding mappings are governed by dgps. We make the inference of the model tractable and scalable by approximating the dgps using random feature expansions and by inferring the resulting model through stochastic variational inference that could exploit distributed and GPU computing. The proposed dgp-ae is able to flexibly model data with mixed-types feature, which is actively investigated in the recent literature (Vergari et al. 2018). Furthermore, the model is easy to implement using automatic differentiation tools, and is characterized by robust training given that, unlike most gp-based models (Dai et al. 2016), it only involves tensor products and no matrix factorizations.
Through a series of experiments, we demonstrated that dgp-ae s achieve competitive results against state-of-the-art novelty detection methods and dnn-based novelty detection methods. Crucially, dgp-ae s achieve these performance with a practical learning method, making deep probabilistic modeling as an attractive model for general novelty detection tasks. The encoded latent representation is probabilistic and it yields uncertainty that can be used to turn the proposed autoencoder into a generative model; we leave this investigation for future work, as well as the possibility to make use of dgps to model the mappings in variational autoencoders.
Notes
References
Abadi, M., Agarwal, A., Barham, P., Brevdo, E., Chen, Z., Citro, C., Corrado, G. S., et al. (2015). TensorFlow: Large-scale machine learning on heterogeneous systems. Software available from tensorflow.org.
Arjovsky, M., Chintala, S., & Bottou, L. (2017). Wasserstein gan. arXiv:1701.07875v2
Bache, K., & Lichman, M. (2013). UCI machine learning repository. Irvine: University of California, School of Information and Computer Sciences. http://archive.ics.uci.edu/ml.
Bradshaw, J., Alexander, & Ghahramani, Z. (2017). Adversarial examples, uncertainty, and transfer testing robustness in Gaussian process hybrid deep networks. arXiv:1707.02476
Bui, T. D., Hernández-Lobato, D., Hernández-Lobato, J. M., Li, Y., & Turner, R. E. (2016). Deep Gaussian Processes for regression using approximate expectation propagation. In M. Balcan & K. Q. Weinberger (Eds.), Proceedings of the 33nd international conference on machine learning, ICML 2016, New York City, NY, USA, June 19–24, 2016, volume 48 of JMLR workshop and conference proceedings (pp. 1472–1481). JMLR.org.
Cho, Y., & Saul, L. K. (2009). Kernel methods for deep learning. In Y. Bengio, D. Schuurmans, J. D. Lafferty, C. K. I. Williams, & A. Culotta (Eds.), Advances in neural information processing systems (Vol. 22, pp. 342–350). Curran Associates, Inc.
Collobert, R., & Weston, J. (2008). A unified architecture for natural language processing: Deep neural networks with multitask learning. In Proceedings of the 25th international conference on machine learning, ICML ’08 (pp. 160–167). New York, NY: ACM.
Cutajar, K., Bonilla, E. V., Michiardi, P., & Filippone, M. (2017). Random feature expansions for Deep Gaussian Processes. In D. Precup & Y. W. Teh (Eds.), Proceedings of the 34th international conference on machine learning, volume 70 of proceedings of machine learning research (pp. 884–893). Sydney: International Convention Centre, PMLR.
Dai, Z., Damianou, A., González, J., & Lawrence, N. (2016). Variationally auto-encoded Deep Gaussian Processes. In Proceedings of the fourth international conference on learning representations (ICLR 2016).
Damianou, A. C., & Lawrence, N. D. (2013). Deep Gaussian Processes. In Proceedings of the sixteenth international conference on artificial intelligence and statistics, AISTATS 2013, Scottsdale, AZ, USA, April 29–May 1, 2013, volume 31 of JMLR proceedings (pp. 207–215). JMLR.org.
Davis, J., & Goadrich, M. (2006). The relationship between precision–recall and ROC curves. In Proceedings of the 23rd international conference on Machine learning, ICML ’06 (pp. 233–240). New York, NY: ACM.
Dereszynski, E. W., & Dietterich, T. G. (2011). Spatiotemporal models for data-anomaly detection in dynamic environmental monitoring campaigns. ACM Transactions on Sensor Networks (TOSN), 8(1), 3.
Dinh, L., Sohl-Dickstein, J., & Bengio, S. (2016). Density estimation using real NVP. arXiv:1605.08803
Domingues, R., Filippone, M., Michiardi, P., & Zouaoui, J. (2018). A comparative evaluation of outlier detection algorithms: Experiments and analyses. Pattern Recognition, 74, 406–421.
Duvenaud, D. K., Rippel, O., Adams, R. P., & Ghahramani, Z. (2014). Avoiding pathologies in very deep networks. In Proceedings of the seventeenth international conference on artificial intelligence and statistics, AISTATS 2014, Reykjavik, Iceland, April 22–25, 2014, volume 33 of JMLR workshop and conference proceedings (pp. 202–210). JMLR.org.
Emmott, A., Das, S., Dietterich, T., Fern, A., & Wong, W.-K. (2016). A meta-analysis of the anomaly detection problem. arXiv:1503.01158v2
Gal, Y., & Ghahramani, Z. (2016). Dropout as a Bayesian approximation: Representing model uncertainty in deep learning. In Proceedings of the 33rd international conference on international conference on machine learning—volume 48, ICML’16 (pp. 1050–1059). JMLR.org.
Gal, Y., Hron, J., & Kendall, A. (2017). Concrete dropout. arXiv:1705.07832
Garca, S., Fernndez, A., Luengo, J., & Herrera, F. (2010). Advanced nonparametric tests for multiple comparisons in the design of experiments in computational intelligence and data mining: Experimental analysis of power. Information Sciences, 180(10), 2044–2064.
Goodfellow, I., Pouget-Abadie, J., Mirza, M., Xu, B., Warde-Farley, D., Ozair, S., Courville, A., & Bengio, Y. (2014) Generative adversarial nets. In Z. Ghahramani, M. Welling, C. Cortes, N. D. Lawrence, & K. Q. Weinberger (Eds.), Advances in neural information processing systems (Vol. 27, pp. 2672–2680). Curran Associates, Inc.
Graves, A. (2011). Practical variational inference for neural networks. In J. Shawe-Taylor, R. S. Zemel, P. L. Bartlett, F. Pereira, & K. Q. Weinberger (Eds.), Advances in neural information processing systems (Vol. 24, pp. 2348–2356). Curran Associates, Inc.
Greensmith, J., Twycross, J., & Aickelin, U. (2006). Dendritic cells for anomaly detection. In IEEE international conference on evolutionary computation, 2006 (pp. 664–671). https://doi.org/10.1109/CEC.2006.1688374.
Hinton, G., Deng, L., Yu, D., Dahl, G. E., Mohamed, A.-R., Jaitly, N., et al. (2012). Deep neural networks for acoustic modeling in speech recognition: The shared views of four research groups. IEEE Signal Processing Magazine, 29(6), 82–97.
Hodge, V., & Austin, J. (2004). A survey of outlier detection methodologies. Artificial Intelligence Review, 22(2), 85–126.
Japkowicz, N., & Stephen, S. (2002). The class imbalance problem: A systematic study. Intelligent Data Analysis, 6(5), 429–449.
Kemmler, M., Rodner, E., Wacker, E.-S., & Denzler, J. (2013). One-class classification with Gaussian processes. Pattern Recognition, 46(12), 3507–3518.
Kim, J., & Scott, C. D. (2012). Robust kernel density estimation. Journal of Machine Learning Research, 13, 2529–2565.
Kingma, D. P., & Welling, M. (2014). Auto-encoding variational Bayes. In Proceedings of the second international conference on learning representations (ICLR 2014).
Krizhevsky, A., Sutskever, I., & Hinton, G. E. (2012). ImageNet classification with deep convolutional neural networks. In Proceedings of the 25th International conference on neural information processing systems, NIPS’12 (pp. 1097–1105). Curran Associates Inc.
LeCun, Y., Bengio, Y., & Hinton, G. (2015). Deep learning. Nature, 521(7553), 436–444.
Liu, F. T., Ting, K. M., & Zhou, Z.-H. (2008). Isolation forest. In Proceedings of the 2008 eighth IEEE international conference on data mining, ICDM ’08 (pp. 413–422). IEEE Computer Society.
Liu, H., Shah, S., & Jiang, W. (2004). On-line outlier detection and data cleaning. Computers & Chemical Engineering, 28(9), 1635–1647.
Maaten, L., & Hinton, G. (2008). Visualizing data using t-SNE. Journal of Machine Learning Research, 9(Nov), 2579–2605.
Markou, M., & Singh, S. (2003). Novelty detection: A review-part 2: Neural network based approaches. Signal Processing, 83(12), 2499–2521.
Neal, R. M. (1996). Bayesian learning for neural networks (lecture notes in statistics) (1st ed.). Berlin: Springer.
Pimentel, M. A., Clifton, D. A., Clifton, L., & Tarassenko, L. (2014). A review of novelty detection. Signal Processing, 99, 215–249.
Pokrajac, D., Lazarevic, A., & Latecki, L. J. (2007). Incremental local outlier detection for data streams. In Computational intelligence and data mining, 2007. CIDM 2007. IEEE symposium on (pp. 504–515). IEEE.
Prastawa, M., Bullitt, E., Ho, S., & Gerig, G. (2004). A brain tumor segmentation framework based on outlier detection. Medical Image Analysis, 8(3), 275–283.
Rasmussen, C. E., & Williams, C. (2006). Gaussian processes for machine learning. Cambridge, MA: MIT Press.
Robbins, H., & Monro, S. (1951). A stochastic approximation method. The Annals of Mathematical Statistics, 22, 400–407.
Srivastava, N., Hinton, G., Krizhevsky, A., Sutskever, I., & Salakhutdinov, R. (2014). Dropout: A simple way to prevent neural networks from overfitting. Journal of Machine Learning Research, 15(1), 1929–1958.
Tipping, M. E., & Bishop, C. M. (1999). Probabilistic principal component analysis. Journal of the Royal Statistical Society: Series B (Statistical Methodology), 61(3), 611–622.
Uria, B., Côté, M.-A., Gregor, K., Murray, I., & Larochelle, H. (2016). Neural autoregressive distribution estimation. Journal of Machine Learning Research, 17(205), 1–37.
Vergari, A., Peharz, R., Di Mauro, N., Molina, A., Kersting, K., & Esposito, F. (2018). Sum-product autoencoding: Encoding and decoding representations using sum-product networks. In Proceedings of the AAAI conference on artificial intelligence (AAAI).
Worden, K., Manson, G., & Fieller, N. R. (2000). Damage detection using outlier analysis. Journal of Sound and Vibration, 229(3), 647–667.
Zimek, A., Schubert, E., & Kriegel, H.-P. (2012). A survey on unsupervised outlier detection in high-dimensional numerical data. Statistical Analysis and Data Mining, 5(5), 363–387.
Acknowledgements
The authors wish to thank the Amadeus Middleware Fraud Detection team directed by Virginie Amar and Jeremie Barlet, led by the product owner Christophe Allexandre and composed of Jean-Blas Imbert, Jiang Wu, Damien Fontanes and Yang Pu for building and labeling the transactions, shared-access and payment-sub datasets. MF gratefully acknowledges support from the AXA Research Fund.
Author information
Authors and Affiliations
Corresponding author
Additional information
Editors: Jesse Davis, Elisa Fromont, Derek Greene, and Bjorn Bringmaan.
Rights and permissions
About this article
Cite this article
Domingues, R., Michiardi, P., Zouaoui, J. et al. Deep Gaussian Process autoencoders for novelty detection. Mach Learn 107, 1363–1383 (2018). https://doi.org/10.1007/s10994-018-5723-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10994-018-5723-3