Abstract
Context
Typical landscape-scale studies comprise the delimitation of landscapes followed by the calculation of one or more landscape metrics. Performing an analysis at multiple spatial scales is often required, occasionally followed by the selection of a particular scale according to the response variable of interest. More complex research goals might require a thorough inspection of landscapes, plus a selection of landscapes that would fulfill certain conditions regarding their landscape metrics. These tasks can usually be programmatically challenging, especially if multiple spatial scales are being analyzed.
Objectives
The R package multilandr builds on several spatial-oriented R packages to provide a toolbox to develop and inspect multi-scale landscapes based on simple spatial inputs.
Methods and results
The package delivers functions to calculate metrics within a multi-scale framework. Also, it provides several utility functions to visualize correlations between metrics, filter landscapes that fulfill certain predefined conditions or select a wide-range gradient of landscapes for a given metric, among other useful tasks. This paper introduces the functionality of multilandr through a step-by-step instruction guide and case studies.
Conclusions
The R package multilandr provides a set of functions to facilitate typical methodological workflow of landscape-scale studies in the R environment, for both beginner and expert R users. It provides the functionality to perform a systematic filtering and selection of landscapes according to a given experimental design. The package is especially programmed to develop multi-scale designs but is also useful for the calculation of metrics of a set of landscapes from any GIS-related project.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
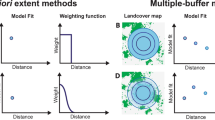
Countless studies on environmental and biological sciences require the delimitation of landscapes. There is not a single definition for landscape (Turner and Gardner 2015), although there is agreement that spatial pattern or heterogeneity and spatial scale are key concepts within the discipline of landscape ecology (Wu 2007, 2013). A landscape can be defined at diverse spatial scales (from a few square meters to hundreds of kilometers) or shaped with different forms (circles, squares or even arbitrarily shaped according to political boundaries). Yet, from a strictly technical point of view, one can simply define a landscape as a buffer area, typically circular, of a fixed radius around a geographical point location (Brennan et al. 2002). This methodological definition of landscape has been widely used among landscape ecology studies for the past years (e.g. Anderle et al. 2023; Faria et al. 2023; Hämäläinen et al. 2023; Parra-Sanchez and Edwards 2024). Selecting landscapes within a study region is a key step for any project aiming to characterize the attributes of a region (e.g. forest amount, forest fragmentation, landscape heterogeneity, etc.) or to assess the effect of certain landscape attributes on a response variable of interest. This is the case, for instance, of studies measuring the relationship between biodiversity (e.g. species richness or abundance of a given species) and the amount of natural habitat in the landscape (e.g. Melo et al. 2017; Galán-Acedo et al. 2018; Verga et al. 2021).
The selection of the spatial scale can influence the results of a study (Wu 2007; Wheatley and Johnson 2009; Jackson and Fahrig 2012, 2015). Ecological responses act at different spatial scales and, if species are involved, the selected scale depends on the biology of the studied species. One common approach to studying the link between ecological processes and scale is to evaluate multiple spatial scales. For circular-shaped landscapes, this can be achieved by generating multiple buffers of increasing radii from the central point of the landscape (Brennan et al. 2002) (Fig. 1). Then, one or more response variables are measured at each landscape of different area, and results can be discussed at the context of multiple spatial scales. Usually, it is desirable to select a scale of effect through some statistical approach, given a set of evaluated spatial scales and a given response variable (Jackson and Fahrig 2012; Huais 2018). The scale of effect would indicate the spatial scale at which the response variable exhibits the strongest relationship with the evaluated landscape attribute, supported by the strongest statistical model (e.g. the model with the highest R2 value or the lowest value for the Akaike Information Criterion) (Huais 2018).
Another important aspect of every landscape-scale study lies in an optimal selection of the set of landscapes that will be evaluated. Depending on the research question and on the proposed experimental design, the geographical locations might be selected to get landscapes that fulfill certain conditions. For example, if the goal of the study is to evaluate the relationship of a given response variable with a landscape metric (e.g. the abundance of pest birds in relation to the amount of forest; Verga et al. 2021), a good experimental design would require that the points are selected in order to get landscapes that comprise a gradient with wide-range values of the landscape metric of interest (i.e. a complete range including low, medium and high values). More complex experimental designs may require gradients of landscapes where certain metrics exhibit low or null correlation, to statistically pull apart the differential effects of distinct landscape metrics. For instance, De Camargo et al. (2018) tested the relationship between avian richness and the number of forest patches (as a measure of habitat fragmentation) but maintaining constant the amount of habitat in the selected landscapes.
Beyond the particularities of each study, the calculation of landscape metrics is a crucial step, and can involve a more complex workflow if the study involves a multi-scale analysis (Fig. 1). Several methods and software are available for the delimitation of landscapes to be studied, as well as for the calculation of metrics. Geographical Information Systems (GIS) such as QGIS (QGIS Development Team 2009) comprise the primary software for these tasks, which allows the preprocessing, analysis and visualization of geographical data, among numerous spatial-related tasks. There is also more specific software oriented to the calculation of diverse landscape metrics, ‘FRAGSTATS’ (McGarigal 1995) being the pioneering software for this task. Specific spatial-oriented packages and scripts are available by several programming languages, such as Python (McClain 2022) or Julia (Hoffimann 2023). Nevertheless, the popularity of the R language (R Core Team 2023) has been increasing in the past decades (Thieme 2018), and is probably the most popular language being used by ecologists nowadays (Lai et al. 2019). Several R packages are currently available for landscape-scale studies (Hesselbarth et al. 2021), such as the package landscapemetrics (Hesselbarth et al. 2019), which provides a tidy workflow for the calculation of numerous landscape metrics from an inputted raster layer.
Although the available R packages provide the necessary framework to conduct landscape-scale studies, this can often be overwhelming for inexperienced R users, or time-consuming for the more experienced ones. Tasks can be especially challenging if the study involves a multi-scale approach, this is, the generation of multiple landscapes at different spatial scales (Fig. 1), and the subsequent calculation of landscape metrics at each spatial scale. This paper introduces the R package multilandr, that builds on several spatial-oriented R packages and comprises a set of functions to process, visualize and analyze spatial data at a landscape-scale. The package provides utility functions to: (i) plot and analyze multi-scale landscapes, (ii) calculate and visualize correlations between metrics, (iii) plot the value of landscape metrics against increasing spatial scales, (iv) filter landscapes that fulfill certain conditions regarding their landscape metrics, (v) select landscapes that comprise gradients with a wide-range of values for a given metric, and (vi) prepare datasets for ulterior statistical tasks.
Software
The R package multilandr provides a toolbox to facilitate a typical workflow of landscape-scale studies. Figure 2 shows a synthesis of the package’s main functionalities. The primary function of the package is mland(), whose output is an R object of class ‘MultiLand’ (Table 1a). Among other inputs, this object is constructed by processing three main user inputs: a raster layer, a spatial points layer and a set of spatial scales. In a previous step, the user might generate a set of points according to certain user-defined rules with the function generate_points(). The object of class ‘MultiLand’ is the primary object of the package, as it comprises all buffers generated around the inputted points given the provided spatial scales, as well as the intersections between the provided raster layers and these buffers (i.e. the actual landscapes). Provided raster layers can represent different land covers (i.e. raster layers with discrete values) or depict continuous variables across the landscape (e.g. climate data, topographic data or other relevant spatial data). This object class serves as the main argument for other functions, including mland_plot() to visualize landscapes and buffers, and mland_overlap() to calculate the degree of overlapping between the different generated buffers.
Workflow of the main R functions provided by the package. Tasks orbit around objects of classes ‘MultiLand’ and ‘MultiLandMetrics’. The first contains all the relevant information and objects regarding landscapes and spatial scales, while the latter contains data related to the calculations of landscape metrics from a ‘MultiLand’ object
In the next step (Fig. 2), the class ‘MultiLand’ serves as the main input for the function mland_metrics(), which allows the calculation of diverse landscape metrics for each landscape at each specified spatial scale (Table 1b). This function mainly relies on the R package landscapemetrics (Hesselbarth et al. 2019), which allows the calculation of diverse landscape metrics, yet the output of this package is then adapted by multilandr for a more straightforward usage for multi-scale approaches. The default output of the function mland_metrics() is an object of class ‘MultiLandMetrics’, which is a mandatory object for other functions of the package. This object contains the data frame with the values of the metrics the user initially required to be calculated, for each landscape of a given site and spatial scale. The object of class ‘MultiLandMetrics’ serves as the main input for other functions programmed to: (i) select landscapes whose metrics present certain values provided by the user with metrics_filter(); (ii) select landscapes that comprise a gradient with wide-range values for a given landscape metric with metrics_gradient(); (iii) perform pairwise metric correlations and plots with metrics_corr() and metrics_plots(), respectively; (iv) generate scalograms, which are plots that relate the value of a given metric in relation to increasing spatial scales (radii) (e.g. Wu 2004); and (iv) prepare the data frame that contains the metric’ values for ulterior visual or statistical analyses with metrics_bind().
In the following section, we present two case studies that exemplify the functionality of the package, inspired by the generation of potential research questions. The first case study exhibits the workflow of a typical landscape-scale study that aim to assess biodiversity in a fragmented landscape, through the definition of an arbitrary landscape gradient. The second case study exhibits the workflow of a more rigorous experimental design, where hundreds of landscapes that fulfill certain conditions are generated. Full arguments of the presented functions can be explored by the user through the help page of each function in the R environment.
Case studies
Case study: exploring the relationship between landscape metrics and biodiversity
Common questions for a typical landscape ecology study focused on evaluating the effects of land use change on biodiversity are: (i) how does the processes of forest loss and fragmentation affect biodiversity? (ii) how does biodiversity respond at different spatial scales? and (iii) is there a spatial scale for which the studied species responds more strongly? To answer these questions, we first need to define a gradient of landscapes, from which one or more landscape metrics can be calculated. Also, research questions (i) and (ii) require performing this analysis at multiple spatial scales. Finally, we also might need to get relevant covariates, occurring across the studied landscapes, that could influence the response variable. Abundant scientific literature has explored these questions through similar methodological approaches (e.g. Melo et al. 2017; Galán-Acedo et al. 2018; Verga et al. 2021; Huais et al. 2024).
For this example, we introduce a potential experimental design geographically located at the southern portion of the Chaco ecoregion in Argentina, where landscapes of different spatial scales are generated from 15 different arbitrary sites. Primary inputs include: (i) raster layers from which landscape metrics will be calculated, and (ii) a vector layer with the geographical location of the sites that will delimit the landscapes to be generated. In this example, the main raster layer comprises a land cover layer from a clipped area of the South American Chaco ecoregion for the year 2000 (Project MapBiomas Chaco 2022). This main raster layer serves as the primary layer for the calculation of traditional landscape metrics from discrete values, as the ones provided by R package landscapemetrics (e.g. percentage of landscape, number of patches, mean patch size of a given raster class, etc.). In addition, we also provide a raster layer of the same extent, containing the mean values of the Normalized Difference Vegetation Index (NDVI) for the year 2000 (Landsat-7 image courtesy of the U.S. Geological Survey). Inputting continuous raster layers allows the calculation of other metrics suitable for continuous data (e.g. mean value or standard deviation). NDVI values are considered as a proxy of vegetation biomass (Rouse et al. 1974), probably influencing our variable of interest (richness of a given group of species), which is relevant in the context of the research questions raised in this case study.
The first steps load the raster and a vector layer with the 15 different geographical points with functions from the package terra (Hijmans 2023).

To easily identify each individual land cover (class) of the main raster layer (named “elchaco”), we assign a meaningful name for each raster class. The main raster layer with discrete values comprises six distinct classes.

Next, we generate a ‘MultiLand’ object with the function mland().

Mandatory inputs include at least one raster layer and a vector layer containing the geographical locations of sites. Main raster layers intended to be used for the calculation of traditional landscape metrics must be inputted in the argument rast_layer (in this example, the object named “elchaco”), whereas other raster layers intended to be used to perform other calculations must be inputted in the argument ext_rast_layer (in this example, the object named “elchaco_ndvi”). Also, we must define the spatial scales from which landscapes will be generated. In this case study, we define the spatial scales as follows: 1000 m until 5000 m, by 1000 m steps. This could be the case, for instance, of a study aiming to find the scale of effect of avian richness in relation to the amount of one or more land covers (e.g. Paton et al. 2019; Mao et al. 2023). Given that each point contained in the points vector layer has an associated name or id for ulterior identification, we provide the name of the attribute inside the points vector layer containing this information, by passing a string through the argument site_ref.
In addition, we input the names of the classes (inside argument class_names) and specify names for the raster layers (rast_names). At last, by setting the argument on_the_fly to FALSE, we are requesting the function to perform the intersections between the buffers (of radii equal to the inputted in argument radii) and the raster layers inputted in arguments rast_layer and/or ext_rast_layer. Thus, actual landscapes will be generated and stored in the output of the function. In the next case study, we show the advantages of generating a ‘MultiLand’ object with the argument on_the_fly equal to TRUE. Calling the generated object by its name provides basic information, including the number of raster layers, points and spatial scales.
An initial aspect we might want to know is the degree (%) of spatial overlapping between the different landscapes, for which the function mland_overlap() will do the task. This simply outputs a two-dimensional matrix with the percentage of overlap between buffers. Next, landscapes can be plotted for a quick visual inspection (Fig. 3), with the function mland_plot().
> mland_plot(ernesdesign, title = "sitename")
Output of the function mland_plot(), showing the plots of each landscape contained in a ‘MultiLand’ object, with their respective assigned names. These are the intersections between a provided raster layer and the specified buffers (from 1000 to 5000 m, by 1000 m steps), around each geographical site
Spatial objects contained inside a ‘MultiLand’ object (points, buffers and intersections between buffers and raster layers) can be exported as GIS data through mland_export_gis() for further investigation or plotting, such as through QGIS (QGIS Development Team 2009). The function will export a zip file containing points, buffers and the intersections between buffers and raster layers (actual landscapes). In addition, the ‘MultiLand’ object can be saved with the function mland_save(), and then loaded in a fresh R session with mland_load().
Once generated the ‘MultiLand’ object, we can calculate several landscape metrics through the function mland_metrics(). In this example, the function will calculate the percentage of the landscape ("pland") and the edge density ("ed") for each class (level = "class") of the main raster layers (raster layers with discrete values depicting different land covers) of ‘MultiLand’. In line with the research questions of this case study, these metrics act as measures of forest cover and forest fragmentation. The calculation of edge density is preferred among other metrics (such as the number of patches) because its value is not scale-dependent, a relevant aspect to consider if we are analyzing multiple spatial scales. Also, for the extra raster layers (argument ext_calc), in this case the function will calculate the mean values of the NDVI.

Some details regarding the inputted parameters need further explanation. On one side, the argument absence_values indicates the value that each class-level metric should acquire when a given class (land cover) is not present in each landscape (e.g. there are no urban areas in a given landscape). Given the nature of one of the metrics being calculated, the percentage of landscape for a given class, the value should equal zero in the absence of this class within each landscape. If this is not clarified, by default the value will be NA (absence of value). On the other side, a list must be provided in argument ext_calc, comprised by as many elements as required metrics, each element consisting in a vector of two internal elements: the first one defines the raster layer (within the extra raster layers provided in argument ext_raster, during the calling of mland() for the generation of an object of class ‘MultiLand’) and the second one the name of the function to be applied to the values of the landscapes for the specified raster layer. The required function can be a function of a loaded package (e.g. "mean" or "sd"), or a user-defined function from the R environment.
Numerous landscape metrics can be calculated, which can be specified through the arguments level, metric, name, type and/or what, the same required by the function calculate_lsm() from the landscapemetrics package (Hesselbarth et al. 2019). Specifications for each argument can be explored through the help page (?mland_metrics). Available landscape metrics can be explored through metrics_list().
By default, mland_metrics() outputs an object of class ‘MultiLandMetrics’. Calling the generated object by its name provides basic information regarding which metrics were calculated, for which layers, classes, points and radii. The most relevant element of this object is the data frame containing the values of the required metrics, which may be exported for further use by the user (Table 2). The data frame is mainly based on the output generated by the function calculate_lsm() from the package landscapemetrics, but rearranged for a multi-scale approach. Each row contains the value of a given metric for a given landscape around a central point from a given raster layer and radius (Table 2). If the required metric is a class-level metric, the class and the name of the class is informed under the columns “class” and “classname”. If the required metric is a patch-level metric, the id of the patch is informed under the column “patch_id”. The function also allows to output this data frame directly, but an object of class ‘MultiLandMetrics’ (the default output) can be passed to other functions of the package (Fig. 2).
A logical next step would be to associate these metrics to a data frame containing sampling data from each individual site (i.e. a data frame containing a column for the site identification and another column with the values of the measured response variable for each site). These metrics include those calculated for the main raster layers (land covers: the percentage of the landscape and edge density for each class) and for the extra raster layers (NDVI values). This can be accomplished through metrics_bind(), which generates a data frame, with the same rows and columns as the data frame with sampling data, plus new columns with the values for each metric. The wide format of this data frame allows it to be passed through a function to fit a statistical model (e.g. through a formula: response_variable ~ metric_1 + metric_2 + … + metric_n) (Huais 2018), which would have the landscape metrics of land covers as main predictor variables, and the NDVI values as relevant covariates.
An experimental design like the exposed here, which assesses several spatial scales, is useful for evaluating the pattern of change in the value of a landscape metric at different spatial scales (Wu 2004). The package provides this functionality through the function metrics_scalogram(), which plots the value of a landscape metric across increasing spatial scales (i.e. radii). Details about this function plus several examples can be explored in supplementary material and in the help page of the function.
Case study: disentangling the differential effects of landscape metrics
Ecologists have engaged some rich discussions regarding the effects of habitat loss and habitat fragmentation on biodiversity (e.g. Fahrig 2013, 2017; Hanski 2015; Fletcher et al. 2018; Fahrig et al. 2019; Saura 2021; Valente et al. 2023). Among various aspects to consider, some authors claim that the effect of habitat loss can be disentangled from the effect of habitat fragmentation (e.g. Fahrig 2017), whereas other have stated that these processes are coupled and cannot be analyzed separately (e.g. Hanski 2015). In any case, one can define habitat fragmentation as a landscape pattern (rather than a process) occurring at a landscape level (usually defined as habitat fragmentation per se) (Fahrig 2017) and define one or more landscape metrics to quantify this pattern (e.g. Melo et al. 2017; De Camargo et al. 2018; Nery et al. 2018). In this context, we could ask the following question: how does habitat loss affect biodiversity when controlling for habitat fragmentation? In addition, if biodiversity is measured within a focal forest patch, one could hypothesize that the size of the patch could influence the response variable (Hanski 2015), so this variable could be controlled by the experimental design.
As a first step, the proposed design would require selecting a gradient of landscapes with nearly fixed values for the area of the forest patches. The package multilandr provides the tools to perform such tasks. First, given a study area, we generate random point locations through the function generate_points(). This function outputs an object of class ‘SpatVector’ (from package terra; Hijmans 2023) with the generated points. As in the first case study, the study area comprises a raster layer with land covers from a clipped area of the South American Chaco ecoregion for the year 2000 (Project MapBiomas Chaco 2022).

By defining the argument approach as "patch", points will be generated inside patches that fulfill the conditions specified in the argument patch_conditions. In the latter, a list of conditions must be provided through the helper function conditions(), each condition in a distinct list. In this example, the first element of the required condition refers to the class of the patches (1, which for the given raster layer is the “forest” class). The second element "area" means that selected forest patches must have an area that ranges between a minimum of 8 hectares (third element) and 12 hectares (fourth element). The second element must be a patch-level metric, and may be any patch-level metric provided by package landscapemetrics (a list of them is accessible through metrics_list()). Besides generating points through the "patch" approach, the function offers a "random" and a "grid" option. Finally, by defining argument "padding" as 2500, we inform the function not to generate points within 2500 m from the margins of the raster.
Then, as in the first example, we generate a ‘MultiLand’ object through mland(). In this case, we set on_the_fly = TRUE to inform the function not to generate in this step the intersections between buffers around sites and the provided raster layers. The advantage of this is that memory will not be occupied with an unnecessary large object, and intersections will be generated when required by the package “on the fly”. For this example, we specify a unique radius of 2000 m.

As in the first case study, we then calculate different landscape metrics through mland_metrics(). For this example, we calculate the percentage of landscape occupied by forest and the number of forest patches. Absence values are set to zero for both metrics, as this should be the value in the case no land cover is present in the landscape (Table 1b).

The function metrics_corr() calculates the correlation between these metrics. Similarly the function metrics_plots() allows to visualize relationships between different metrics, between different classes of the same metrics and between different radii of the same metric and class (Fig. 4).

Output of the function metrics_plots(). Pair plots between the value of different metrics (percentage of landscape -pland- and number of patches -np- for the class “Forest”) and different radii (in this case, only one radius: 2000 m). The abbreviation “r1” for each plot title refers to the raster layer number one. Diagonal plots depict the distribution of data points of the values of the metrics labeled in the columns. For the purpose of the example, those landscapes selected after applying a filter to the number of patches (with metrics_filter()) are colored (in green or purple), and solely in purple those 30 landscapes selected to be part of the gradient (with metric_gradient()). See the main text for details
Based on the research question, we need to control for forest fragmentation. A common measure for this landscape pattern is the number of forest patches within a given landscape (e.g. De Camargo et al. 2018). This number should be relatively fixed, this is, the number of forest patches should be somehow similar across all landscapes. A rapid inspection of Fig. 4 shows a wide range of values for the number of patches within these landscapes, from 1 to almost 60 patches, which is clearly too large if we aim to control for habitat fragmentation. A precise selection of landscapes can be done through the function metrics_filter(), by inputting an object of class ‘MultiLandMetrics’ generated with mland_metrics().

Here, the argument conditions is generated through the helper function of the same name, and each condition must be defined as a list with the following elements in the expressed order: raster layer, class, radii, metric to be considered in this condition, and the minimum and maximum values for the specified metric. In this case, we specify that all selected landscapes of radius 2000 m for all raster layers (by setting the first element as NA, although in this example there is only one raster layer) must present between 10 and 15 forest patches. As expected, only a portion of the sites fulfill the required condition (totaling 136, ~ 22% of the original number of sites). In Fig. 4, we can see that for the selected points (colored, either in green or purple), the number of forest patches remain relatively fixed (between 10 and 15 patches), while the forest amount ("pland") presents a wide range of values, between ~ 1 to ~ 65%.
Finally, moving a bit away from the main research question of this case study, we could assume that a researcher need to select landscapes for field sampling. In this case, a logical next step would be to select a realistic number of these landscapes to sample. This set of landscapes would comprise a gradient for the percentage of forest. For the gradient to be robust, this should present a wide range of values for this landscape metric, covering a good range of intermediate values between minimum and maximum values. This task can be performed with the function metrics_gradient(), as follows:

Here, we are requesting a gradient of 30 points (argument n) for the metric "pland" and class "Forest" (from raster layer 1 and radius 2000 m). The algorithm will select 30 landscapes that present values for the specified landscape metric that are the closest to a set of numbers or cutpoints. By default, these cutpoints will be calculated as a range of values equally distributed between the minimum and maximum value for the specified metric. Alternatively, to provide more flexibility, the function allows the user to manually specify these cutpoints (see more details in ?metrics_gradient).
In Fig. 4, we can see in purple those selected landscapes that are part of the subset of landscapes previously filtered with metrics_filter(). Overall, after the series of applied filters, we have obtained a gradient of a practical number of landscapes where: (i) central points are located in patches of size between 8 and 12 ha (after calling generate_points()), (ii) the number of forest patches within each landscape ranges between 10 and 15 (after calling metrics_filter()), and (iii) the percentage of forest ranges between absolute minimum and maximum values, with a well-distributed range of intermediate values (after calling metrics_gradient()).
Discussion
The R package multilandr provides the framework to analyze, visualize and process spatial data, specially oriented to develop the first steps to conduct any landscape-scale study. As exemplified in the first case study, the compartmentalization of spatial data in specially programmed object’s classes provides the user with a full control of the whole process of analysis. There is a general awareness in the landscape ecology community that spatial scale can influence the results of a study, and examining multiple spatial scales is preferable to a single-scale approach (Wu 2007). Multi-scale approaches allow to evaluate how landscape attributes or given ecological responses vary across different spatial scales (Wu 2004; Jackson and Fahrig 2015). In addition, assessing several spatial scales is commonly a preliminary step to select a proper scale for a given species or group of species (Jackson and Fahrig 2012, 2015; Wheatley and Johnson 2009). The package provides the necessary tools for multi-scale experimental designs and facilitates the calculation of landscape metrics in this context. It’s worth noting that the values of many landscape metrics are scale-dependent (e.g. the number of patches or the total area of a land cover). The user should carefully consider this issue in multi-scale approaches, as it could result in landscapes of different sizes not being comparable for such metrics. Besides, a quick visual inspection can be performed through the plotting tool. Alternatively, preprocessed spatial data can be exported as GIS data for a more exhaustive examination through a GIS software.
As highlighted in the second case study, the package provides utility functions to perform an optimal selection of landscapes that would fulfill certain conditions regarding the values of their landscape metrics, according to the research interest of the user. This is the case of studies that, for instance, aim to statistically disentangle differential effects of landscape metrics on biodiversity (e.g. Melo et al. 2017; De Camargo et al. 2018; Verga et al. 2021). After the calculation of metrics, the user can perform a precise filtering of landscapes that meet certain conditions. An additional filtering can be made, through the selection of a small but widely ranged gradient of landscapes. This would be the case, for instance, of an experimental design that requires field sampling, but the gradient of potential sampled landscapes must be restricted to reach more realistic number. These are common tasks for experimental designs conducted at a landscape-scale, and multilandr provide the right tools to perform a systematic selection of suitable landscapes according to the design requirements.
The structure of the package, where the functions orbit around classes ‘MultiLand’ and ‘MultiLandMetrics’, facilitate the expansion of functionality of the already available functions, as well as the programming of new ones. For example, new functions related to the selection of a set of landscapes might be useful, such as one that would select landscapes that fulfill more than one attribute at the same time, probably through the application of statistical matching techniques. Besides, the package performs well with both small and large spatial datasets in a regular computer (see Table 3 for the time to run each process in both case studies). However, increasing the speed of some processes for large datasets (e.g. processing thousands of points or large raster layers) might be possible through parallelization. This functionality will be available in further updates of the package.
In conclusion, the presented R package provides a set of useful tools for any landscape-scale study involving an initial delimitation of landscapes followed by the calculation of metrics. This could be the case of studies evaluating relevant landscape ecology questions, but the package is also useful for any study involving GIS-related tasks at a landscape scale. The package also provide functionality to perform a thorough inspection and selection of landscapes that fulfill certain conditions. For those inexperienced R users, these tasks can be overwhelming, or time-consuming for the more experienced ones, and package multilandr addresses this need with a clean framework.
Data availability
No datasets were generated or analysed during the current study.
Software availability
The package can be installed in the R environment from the GitHub repository through the following code: remotes::install_github(“phuais/multilandr”). The code of the case studies is also provided as supplementary material. All the objects being loaded and called for the case studies, as well as help pages and other examples, are available in the R environment after the installation of the package. A manual describing each function in detail can be downloaded from the GitHub repository.
References
Anderle M, Brambilla M, Hilpold A, Matabishi JG, Paniccia C, Rocchini D, Rossin J, Tasser E, Torresani M, Tappeiner U, Seeber J (2023) Habitat heterogeneity promotes bird diversity in agricultural landscapes: insights from remote sensing data. Basic Appl Ecol 70:38–49
Brennan JM, Bender DJ, Contreras TA, Fahrig L (2002) Focal patch landscape studies for wildlife management: optimizing sampling effort across scales. In: Integrating landscape ecology into natural resource management, Cambridge University Press, pp 68–91
De Camargo RX, Boucher-Lalonde V, Currie DJ (2018) At the landscape level, birds respond strongly to habitat amount but weakly to fragmentation. Divers Distrib 24(5):629–639
Fahrig L (2013) Rethinking patch size and isolation effects: the habitat amount hypothesis. J Biogeogr 40(9):1649–1663
Fahrig L (2017) Ecological responses to habitat fragmentation per se. Annu Rev Ecol Evol Syst 48:1–23
Fahrig L, Arroyo-Rodríguez V, Bennett JR, Boucher-Lalonde V, Cazetta E, Currie DJ, Eigenbrod F, Ford AT, Harrison SP, Jaeger JA, Koper N, Martin AE, Martin J-L, Metzger JP, Morrison P, Rhodes JR, Saunders DA, Simberloff D, Smith AC, Tischendorf L, Vellend M, Watling JI (2019) Is habitat fragmentation bad for biodiversity? Biol Conserv 230:179–186
Faria D, Morante-Filho JC, Baumgarten J, Bovendorp RS, Cazetta E, Gaiotto FA, Mariano-Neto E, Mielke MS, Pessoa MS, Rocha-Santos L, Santos AS, Benchimol M (2023) The breakdown of ecosystem functionality driven by deforestation in a global biodiversity hotspot. Biol Conserv 283:110126
Fletcher RJ, Didham RK, Banks-Leite C, Barlow J, Ewers RM, Rosindell J, Holt RD, Gonzalez A, Pardini R, Damschen EI, Melo FP, Ries L, Prevedello JA, Tscharntke T, Laurance WF, Lovejoy T, Haddad NM (2018) Is habitat fragmentation good for biodiversity? Biol Conserv 226:9–15. https://doi.org/10.1016/j.biocon.2018.07.022
Galán-Acedo C, Arroyo-Rodríguez V, Estrada A, Ramos-Fernández G (2018) Drivers of the spatial scale that best predict primate responses to landscape structure. Ecography 41(12):2027–2037
Hämäläinen A, Fahrig L, Strengbom J, Ranius T (2023) Effective management for deadwood-dependent lichen diversity requires landscape-scale habitat protection. J Appl Ecol 60(8):1597–1606
Hanski I (2015) Habitat fragmentation and species richness. J Biogeogr 42(5):989–993
Hesselbarth MH, Sciaini M, With KA, Wiegand K, Nowosad J (2019) landscapemetrics: an open-source R tool to calculate landscape metrics. Ecography 42(10):1648–1657
Hesselbarth MH, Nowosad J, Signer J, Graham LJ (2021) Open-source tools in R for landscape ecology. Curr Landsc Ecol Rep 6(3):97–111
Hijmans R (2023) terra: spatial data analysis. R package version 1.7-29. https://CRAN.R-project.org/package=terra
Hoffimann J (2023) Geospatial data science with Julia. https://juliaearth.github.io/geospatial-data-science-with-julia
Huais PY (2018) multifit: an R function for multi-scale analysis in landscape ecology. Landsc Ecol 33:1023–1028
Huais PY, Grilli G, Galetto L (2024) Impacts of mass flowering soybean on bee visitation rates of forest plants at different spatial scales. Landsc Urban Plan 247:105055
Jackson HB, Fahrig L (2012) What size is a biologically relevant landscape? Landsc Ecol 27:929–941
Jackson HB, Fahrig L (2015) Are ecologists conducting research at the optimal scale? Glob Ecol Biogeogr 24(1):52–63
Lai J, Lortie CJ, Muenchen RA, Yang J, Ma K (2019) Evaluating the popularity of R in ecology. Ecosphere 10(1):e02567
Mao Q, Wu Z, Deng Y, Sun J, Bai H, Gong L, Jiang Z (2023) Assessing the “Scale of Effect”: the impact of multi-scale landscape characteristics on urban bird species taxonomic and functional diversity. Diversity 15(8):943
McClain BP (2022) Python for geospatial data analysis. O’Reilly Media Inc, Sebastopol
McGarigal K (1995) FRAGSTATS: spatial pattern analysis program for quantifying landscape structure, vol 351. US Department of Agriculture, Forest Service, Pacific Northwest Research Station, Portland
Melo GL, Sponchiado J, Cáceres NC, Fahrig L (2017) Testing the habitat amount hypothesis for South American small mammals. Biol Conserv 209:304–314
Nery LS, Takata JT, De Camargo BB, Chaves AM, Ferreira PA, Boscolo D (2018) Bee diversity responses to forest and open areas in heterogeneous Atlantic Forest. Sociobiology 65(4):686–695
Parra-Sanchez E, Edwards DP (2024) Spatial extent predicts Andean epiphyte biodiversity responses to habitat loss and fragmentation across human-modified landscapes. J Biogeogr. https://doi.org/10.1111/jbi.14819
Paton GD, Shoffner AV, Wilson AM, Gagné SA (2019) The traits that predict the magnitude and spatial scale of forest bird responses to urbanization intensity. PLoS ONE 14(7):e0220120
Project MapBiomas Chaco – Collection 4.0 of annual land cover and land use maps, accessed during July 2022 through the following link: https://chaco.mapbiomas.org/
QGIS Development Team (2009) QGIS geographic information system. Open Source Geospatial Foundation Project. http://qgis.osgeo.org
R Core Team (2023) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Rouse Jr JW, Haas RH, Deering DW, Schell JA, Harlan JC (1974) Monitoring the vernal advancement and retrogradation (green wave effect) of natural vegetation (No. E75-10354)
Saura S (2021) The Habitat Amount Hypothesis implies negative effects of habitat fragmentation on species richness. J Biogeogr 48(1):11–22
Thieme N (2018) R generation. Significance 15(4):14–19
Turner M, Gardner R (2015) Landscape ecology in theory and practice: Pattern and process, second edition. https://doi.org/10.1007/978-1-4939-2794-4
Valente JJ, Gannon DG, Hightower J, Kim H, Leimberger KG, Macedo R, Rousseau JS, Weldy MJ, Zitomer RA, Fahrig L, Fletcher RJ Jr, Wu J, Betts MG (2023) Toward conciliation in the habitat fragmentation and biodiversity debate. Landsc Ecol 38(11):2717–2730
Verga EG, Huais PY, Herrero ML (2021) Population responses of pest birds across a forest cover gradient in the Chaco ecosystem. For Ecol Manag 491:119174
Wheatley M, Johnson C (2009) Factors limiting our understanding of ecological scale. Ecol Complex 6(2):150–159
Wu J (2004) Effects of changing scale on landscape pattern analysis: scaling relations. Landsc Ecol 19:125–138
Wu J (2007) Scale and scaling: a cross-disciplinary perspective. In: Key topics in landscape ecology. Cambridge University Press, pp 115–142
Wu J (2013) Key concepts and research topics in landscape ecology revisited: 30 years after the Allerton Park workshop. Landsc Ecol 28:1–11
Acknowledgements
I thank Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) and Universidad Nacional de Córdoba (UNC) for financial support, only possible with sustainable public policies for science. A special thanks to the compromised scientist, ecologist and ornithologist, but above all an endearing friend Ernesto G. Verga, simply “Ernes”. The name “ernesdesign” for the ‘MultiLand’ object created for the first case study of the manuscript was in his honor, as I think it would reflect a typical experimental design from his research in “El Chaco”. You will forever live in our memory.
Funding
The author declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
Pablo Yair Huais: conceptualization, methodology, software, formal analysis, investigation, resources, data curation, writing—original draft, writing—review & editing, visualization, supervision, project administration.
Corresponding author
Ethics declarations
Competing interests
The author has no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Huais, P.Y. multilandr: an R package for multi-scale landscape analysis. Landsc Ecol 39, 140 (2024). https://doi.org/10.1007/s10980-024-01930-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10980-024-01930-z