Abstract
Context
Functional connectivity across fragmented habitat patches is essential for the conservation of animal populations in humanized landscapes. Given their low dispersal capacity, amphibians in the Mediterranean region are threatened by habitat fragmentation and loss due to changes in land use, including agricultural intensification.
Objectives
We assessed patterns of functional connectivity of a Near Threatened Mediterranean amphibian, the sharp ribbed newt (Pleurodeles waltl), in an agricultural landscape matrix in NW Spain subject to different intensification regimes.
Methods
We sampled newts in 17 ponds embedded in a terrestrial habitat matrix dominated by agricultural land uses. Genome-wide molecular markers (1390 SNPs) were used to assess patterns of genetic diversity and gene flow among ponds. We tested the role of landscape features on functional connectivity using isolation by resistance models incorporating information on Normalized Difference Vegetation Index (NDVI) data.
Results
We found low levels of genetic diversity in all sampled populations. Global FST estimates and cluster analyses revealed shallow but significant genetic structure in the study area, with NDVI-based resistance models showing that open areas (rainfed crops and grasslands) offer lower resistance to gene flow and thus promote functional connectivity among demes.
Conclusions
Our study highlights the important role of landscape features, such as open areas resulting from traditional rainfed agriculture, in promoting functional connectivity between amphibian populations in Mediterranean agrosystems. Conservation policies must adopt a functional network strategy and protect groups of inter-connected temporary ponds across the traditional agricultural matrix to efficiently preserve their associated biotic communities.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Habitat destruction and fragmentation are among the main drivers of global biodiversity loss (Newbold et al. 2015; Fardila et al. 2017). Fragmentation causes deleterious effects in spatially-structured populations resulting from decreased connectivity (Haddad et al. 2015), which can be counteracted via demographic rescue if connectivity is restored (Córdova-Lepe et al. 2018). Functional connectivity has been defined as the actual movement of individuals among local populations through the landscape matrix and which successfully reproduce in the receiving populations (Taylor 2006). As such, functional connectivity can be measured by the amount of gene flow (Storfer et al. 2010). When functional connectivity is low, demographic isolation results in reduced population viability, especially in small populations, because of inbreeding and genetic drift (Spurgin and Gage 2019). Therefore, conservation efforts should be directed to assess patterns of inter-patch connectivity, including the identification of suitable habitat corridors and features and configurations of the landscape matrix that facilitate dispersal and prevent the isolation of animal populations.
Agriculture is one of the main drivers of habitat loss and fragmentation, especially in temperate and Mediterranean areas, where mostly primary, pristine habitats are lost (Foley et al. 2005). Traditional agricultural landscapes are compositionally heterogeneous, with different crop types and interspersed patches of natural habitats. However, modern agriculture often tends to promote monocultures to increase yields, resulting in landscape homogenization and biodiversity loss (Benton et al. 2003; Fahrig et al. 2011). For species inhabiting habitat patches within an agricultural matrix, the composition and configuration of croplands within the matrix, as well as the use of agrochemicals and the presence of alien invasive species, represent the main factors potentially affecting the amount of habitat use and inter-patch dispersal, and thus the vulnerability of local populations to extinction due to stochastic events. Reduced connectivity (i.e., gene flow) between populations leads to a concomitant loss of genetic diversity, including beneficial alleles, due to inbreeding and genetic drift (Halverson et al. 2006). In this context, small populations will be more rapidly depleted of genetic diversity, which can be assessed by calculating their effective population size (Ne), an important conservation parameter defined as the size of an ‘ideal’ population that experiences the same rate of change of allele frequencies or heterozygosity as the target population (Wright 1931). Genomic studies at the landscape level can provide estimates of effective population sizes and patterns of gene flow among demes, identifying smaller and/or isolated populations of conservation concern and landscape features promoting or restricting functional connectivity (Taylor 2006; Smith et al. 2009; Marchi et al. 2013).
Pond-breeding amphibians are suitable model systems in landscape genomics studies because their populations are naturally spatially structured due to the discontinuous presence of freshwater breeding habitats in the landscape. Patterns of inter-deme connectivity are shaped by features of the terrestrial habitat and their interaction with dispersal capacity, which is generally assumed to be low but is unknown for most species (Vos et al. 2007; Youngquist et al. 2017; Cayuela et al. 2020). Amphibians in agricultural areas in the Mediterranean region mostly use temporary ponds as preferred breeding sites. The increase in the transformation of rainfed into irrigated crops, as well as the continuous efforts to concentrate crops in larger plots in the agricultural landscape, are favoring the isolation of these wetlands and their biotic communities, potentially compromising their viability in the long term (Beja and Alcazar 2003; Ficetola and De Bernardi 2004; Ferreira and Beja 2013).
Agricultural intensification also leads to the loss and degradation of terrestrial and aquatic habitats used by adult amphibians for both foraging and dispersal, such as non-cropped patches, streams, temporally flooded ditches or traditional irrigation channels (Gray et al. 2004). Despite the potential for agricultural intensification to disrupt connectivity among amphibian demes, few studies have used genomic tools to assess patterns of gene flow in these systems in the Mediterranean region. Wetlands in this region are smaller and more temporary and isolated than those in northern Europe (Bolle et al. 2003; Bekioglu et al. 2007). These differences may impact patterns of amphibian abundance, dispersal and survival, especially in less mobile species like urodeles (salamanders and newts). In particular, small, more isolated populations tend to harbor lower levels of local genetic diversity and display high genetic differentiation among demes at the landscape level, which makes them more sensitive to further habitat fragmentation and loss.
We assessed the influence of agricultural intensification on functional connectivity in an amphibian model, the sharp ribbed newt Pleurodeles waltl Michahelles, 1830. This Ibero-Maghrebian endemic is the largest salamandrid newt in the world and is listed as Near Threatened (NT) by the International Union for the Conservation of Nature (IUCN) because of inferred declines associated with widespread habitat loss and the negative effects of invasive species (Beja et al. 2009). Adult newts are mostly aquatic but can disperse among ponds through the terrestrial habitat matrix during humid or rainy nights (Salvador 2015), although these dispersal events seem to be very rare (Fernández de Larrea et al. 2021; Reyes-Moya et al. 2022). Capture-mark-recapture studies have documented low dispersal rates between breeding sites located up to 700 m apart (Gutiérrez-Rodríguez et al. 2017a; Fernández de Larrea et al. 2021; Reyes-Moya et al. 2022) and found evidence of genetic structure at small spatial scales in natural landscapes (Gutiérrez-Rodríguez et al. 2017c), suggesting the species is highly sensitive to habitat fragmentation. Therefore, we expected to find stronger genetic structure among demes in our study area, where changes in the landscape matrix caused by agricultural intensification are hypothesized to result in reduced dispersal and gene flow among demes, especially considering the low dispersal potential of the species.
We developed a genome-wide panel of molecular markers (single nucleotide polymorphisms, SNPs) to characterize genetic diversity and fine-scale patterns of connectivity among 17 breeding populations (demes) of P. waltl in an agricultural area and explored the relationship between genetic structure and landscape habitat features using Normalized Difference Vegetation Index (NDVI) data. Specifically, we tested the effect on connectivity of land uses related to agricultural intensification vs natural habitat patches, including forested areas and vegetated streams. We discuss the implications of our results for the conservation and management of amphibian populations in agricultural areas in the Mediterranean region.
Materials and methods
Sampling
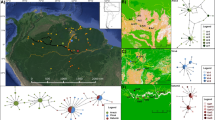
The study area (Fig. 1) is located in southeast León province (Spain), part of the historical region of “Tierra de Campos” in the Iberian northern plateau, between the Esla and Cea rivers, in the Duero basin. The climate is Mediterranean with continental influence. Landscape cover is mostly composed of cereal rainfed crops, with some amount of natural vegetation comprising oak forests (Quercus pyrenaica) and grasslands. The amount of natural vegetation cover increases northwards, and irrigated crops, mostly corn, are more frequent on the western part of the area. The lithological features of this area favor the formation of numerous water bodies, mostly temporary ponds (Fernández Aláez et al. 1999).
Map of the study area showing main land cover types and the location of sampled ponds. Landscape cover data was obtained from the database of “Mapa de Cultivos y Superficies Naturales de Castilla y León” (MCSNCyL, Junta de Castilla y León). The main rivers of Esla (west) and Cea (East) are marked in red
The study area represents the northern range limit for P. waltl, which was found to be widespread in the area during our surveys. We sampled 7–14 adult newts from 17 ponds (total sample size: 185 individuals) along a north–south gradient (Fig. 1; Table S1). While modest, these sample sizes (N) have been proven to be sufficient in other amphibian studies using SNP markers (McCartney-Melstad et al. 2018; Rödin-Mörch et al. 2021). Sampled ponds can be grouped in two major wetland complexes, Payuelos in the north (ponds 1–13), and Oteros in the south (ponds 15, 16 and 17), with one pond in an intermediate position (pond 14). We selected ponds where P. waltl was locally abundant, and that were located in habitats representing the three main land uses in the study area (i.e., rainfed crops, irrigated crops, and natural areas). Distances among ponds ranged from 1 to 35 km (Table S2). Adult newts were captured in their aquatic stage during nocturnal surveys, using funnel traps or by hand, and released immediately after tissue sampling (toe clips, which were preserved in 100% ethanol prior to DNA extraction). All experimental protocols were approved by the regional authority (Consejería de Medio Ambiente, Junta de Castilla y León; permit code AUES/CYL/693/2019).
DNA extraction, SNP calling and genotyping
DNA was extracted from tissue samples using the DNeasy Blood and Tissue kit (Qiagen), following the standard protocol provided by the manufacturer and using RNase. Prior to genomic analyses, we performed a preliminary test of polymorphism using 18 microsatellite markers used in previous studies (e.g., Gutiérrez-Rodríguez et al. 2017b) to genotype 93 individuals from five ponds. This preliminary screening showed a large proportion of monomorphic markers in the sample (9/18, 50%), with no evidence of genetic differentiation among ponds based on Structure runs under different values of K (i.e., genetic clusters).
The 2b-RAD protocol (Wang et al. 2012) was used to obtain a reliable SNP collection across the entire P. waltl genome, following the protocol of Maroso et al. (2019), treating DNA with the 2b-type restriction enzyme AlfI. DNA from the 185 sampled individuals was pooled in three genomic libraries (60–65 individuals each) for sequencing in an Illumina NextSeq500 System. Reads were cleaned using the process_radtags function in the software STACKS v2.4 (Catchen et al. 2013), retaining reads with a size of 36 bp (the expected length of the fragments after digestion with AlfI) and with Phred quality scores > 20 in at least 75% of nucleotides. Reads where the AlfI recognition site was missing were removed. After cleaning, sequences were aligned to the P. waltl genome (Elewa et al. 2017) using the software BOWTIE v1.2 (Langmead et al. 2009) and allowing a maximum of two mismatches (-v 2 option in BOWTIE). Due to the large size of this genome (≈ 20 Gb), fragments containing the AlfI recognition site were extracted using the script ExtractSites.pl from E. Meyer’s lab (Oregon University, USA), prior to alignment. RADtags matching against different regions of the genome were removed to filter out potential paralogs (-m 1 option in BOWTIE). Aligned loci were processed with the STACKS module gstacks with default parameters (model marukilow and alpha threshold 0.05). Then, the populations module was used without any filtering parameter to obtain an initial SNP catalogue. Finally, the software PLINK v1.9 (Chang et al. 2015) was used to implement the following filtering steps and obtain the final catalogue of SNPs: (i) genotyped in at least 75% of individuals (flag –geno 0.25 in PLINK), (ii) minimum allele count (MAC) of 3 (–mac 3 in PLINK), and (iii) consistent with Hardy–Weinberg (HW) equilibrium (p-value > 0.05) in at least 75% of the demes (–hwe 0.05 in each deme and manual editing). Loci not passing these filters were removed prior to downstream analyses.
Identification of outlier loci
SNPs linked to genomic regions under natural selection may bias inferences of neutral genetic structure. Therefore, we used two complementary approaches to identify loci putatively under selection (i.e., outlier loci). First, we applied the Bayesian FST -based method used by BAYESCAN v2.01 (Foll and Gaggiotti 2008), with default parameters (i.e., 20 pilot runs; prior odds value of 10; 100,000 iterations; burn-in of 50,000 iterations and a sample size of 5000). Loci with a False Discovery Rate (FDR) lower than 5% were considered as outliers. Second, we used the R package PCADAPT v4.0 (Luu et al. 2017; Prive et al. 2020), which implements a principal components-based method not requiring a priori population assignment. This method renders low false-positive rates by using individual information. We selected the optimal number of principal components (PC) with the “chooseK” option. For outlier identification we used the q-values, again using as a cut-off a FDR of 5% (q-value < 0.05). We considered as outliers the loci identified by at least one of the two methodologies applied.
Genetic diversity and population structure
Basic population genetics statistics, including the mean number of alleles per locus (Na), observed heterozygosity (HO), expected heterozygosity (HE), and the inbreeding coefficient (FIS), were calculated for each sampled pond (breeding demes) using the software GENEPOP v4.7 (Rousset 2008), ARLEQUIN v3.5 (Excoffier and Lischer 2010), and R package diveRsity (Keenan et al. 2013). In addition to PLINK filtering, conformance to Hardy–Weinberg (HW) expectations for each deme was also assessed with the probability test implemented in GENEPOP. The effective population size (Ne) for each deme was estimated using the linkage disequilibrium method implemented in NeESTIMATOR v2.1 (Do et al. 2014), using the default threshold values for allele frequencies (PCrits) of 0.01, 0.02 and 0.05. Global and pairwise population differentiation (FST) values were calculated using the R package StAMPP (Pembleton et al. 2013), with 10,000 bootstrap replicates to assess their significance, and applying the Bonferroni correction for multiple testing to p-values.
For population structure analyses, we excluded outlier loci (neutral SNP dataset, see Results), but we also report results based on the full SNP catalogue (full SNP dataset, see Supplemental Material). The Bayesian clustering method implemented in STRUCTURE v2.3.4 (Pritchard et al. 2000) was used to assess the number of genetically homogeneous population units (K). For each K value (from one to 17), five replicates were run using an admixture model with correlated allele frequencies, with a burn-in of 50,000 steps and 200,000 post-burn-in iteration steps. Due to the weak structure found, we incorporated information about sampling locations by using the LOCPRIOR option. Results from the STRUCTURE runs were processed with the software STRUCTURE HARVESTER v0.6 (Earl and Von Holdt 2012) to infer the optimal value of K based on the ΔK method (Evanno et al. 2005). The software CLUMPAK (Kopelman et al. 2015) was used for graphical representation of individual cluster assignment probabilities across K values. As an alternative and complementary description of genetic structure, we also performed a discriminant analysis of principal components (DAPC) with the R package ADEGENET (Jombart 2008). Genotypes were transformed via principal components (PCA) and the Bayesian information criterion (BIC) was used to find the optimal number of clusters via the k-means procedure (function find.clusters), retaining 60 principal components (PCs) for the analysis, less than a third of the number of individuals (N/3), as higher numbers of PCs can render membership assignment probabilities unstable (Jombart 2012). DAPC ordination analyses were run for all individuals separately and for the optimal number of clusters returned by the program.
We also ran spatially explicit analyses of population structure in TESS3 (Caye et al. 2016), as implemented in the R package tess3r. The clustering algorithm implemented in TESS uses genetic and geographic data simultaneously, providing better results than non-spatial clustering algorithms when genetic divergence among populations is low. We ran the algorithm to estimate ancestry coefficients for values of K from 1 to 17 with 200 replicates for each. The optimal value of K was assessed using cross-validation scores.
Landscape genomic analyses
Isolation by distance (IBD) was tested by assessing the correlation between the matrices of geographical Euclidean distances between ponds (Table S2) and genetic distances (FST) with a Mantel test, using the mantel function in the R package VEGAN (Oksanen et al. 2017). To test for IBD, we applied the correction of Rousset (1997) for a two-dimensional geographic distribution, using FST/(1 − FST) and the logarithm of geographic distances. IBD tests were carried out using the neutral SNP dataset, but also for the full SNP dataset. The proportion of shared alleles (DPS), as calculated by the R package GRAPH4LG, was also used as a finer and complementary measure of genetic distance to test for IBD.
Due to the semi-aquatic biology of the focal species, and the lack of other major topographical features in the study area, we focused on the potential influence of agricultural vs. forested areas on gene flow and developed a model of landscape resistance between sampled ponds. We obtained values of the Normalized Difference Vegetation Index (NDVI) for the study area, a parameter related to vegetation cover and wetness and which has been identified as a predictor of gene flow in landscape genetics studies involving amphibians (Antunes et al. 2018; Sinai et al. 2019; Velo-Antón et al. 2021), including P. waltl (Gutiérrez-Rodríguez et al. 2017c). A temporal series of NDVI values covering the last 20 years (2002–2022) was used to incorporate information of land use changes. The NDVI time series was obtained from the MOD13Q1 product (version 6.1) based on the MODIS Terra satellite and downloaded from the Earth Data website (www.earthdata.nasa.gov). A harmonic regression to produce a set of five coefficients representing different seasonalities in the NDVI time series (Estrada-Peña et al. 2014) was utilized. The linear combination of these coefficients was optimized with Genetic Algorithms with 500 iterations and values between − 10 to 10, to derive a model of landscape resistance that maximizes connectivity. A Generalized Least Squares (GLS) model between pairwise genetic dissimilarities and the pairwise resistance distances obtained from the optimized resistance surface was then applied. A correlation structure was used based on the maximum likelihood populations effect model (MLPE) to control for the lack of independence resulting from using pairwise distances. Three different models of isolation (Isolation by Distance, Isolation by Resistance, and a model incorporating both predictors) were tested using genetic and distance matrices. These models hypothesize that geographic distances, landscape factors (as described by NDVI values), or their combination, explain genetic distances among populations, respectively. Models were ranked according to the Akaike Information Criterion (AIC) and compared with a null model. All landscape genetic analyses were performed in R with packages RASTER, GDISTANCE, GA and CORMLPE. For landscape genomics analyses, genetic distance matrices were calculated with the neutral SNP dataset, but results obtained with the full SNP dataset are also presented for reference in the Supplemental Material.
Results
Sampling and sequencing
The final dataset contained 184 individuals, for which the populations module from STACKS returned an initial catalogue of 198,897 variable sites (i.e., SNPs). After filtering, a final list of 1528 highly reliable SNPs was retained, with a mean genotyping rate of 151 genotyped individuals per locus (range 139–182, see Tables S8–S10), and a mean percentage of missing genotyped loci per individual of 17.9% (range 4.5–49%).
Identification of outlier loci
Three loci were identified as outliers by Bayescan, and 138 loci were identified by PCADAPT (Table S11). All of them were inferred to be under divergent selection. The three outlier loci identified by Bayescan were included in the 138 loci set identified by PCADAPT; therefore, these markers were removed from downstream analyses, resulting in a neutral SNP dataset of 1390 markers.
Genetic diversity
We found very low values of genetic diversity across all demes (Tables 1, S3), with mean observed heterozygosity (HO) = 0.138 and mean expected heterozygosity (HE) = 0.106 in the neutral SNP dataset. The mean number of alleles per locus (Na) was also low (1.37). Genetic diversity (HE), using the neutral dataset, was not correlated with sample size (R2 = 0.022; p-value = 0.571), and the least diverse deme was that with the largest sample size (Pond 11-Cimera, N = 14). Global FIS values for all demes were negative (i.e., excess of heterozygotes); however, these demes were in accordance with HW expectations, with negative FIS values probably resulting from the low number of individuals analyzed in each deme and/or small effective population sizes (but see below). Diversity values obtained with the complete dataset were similar to those in the neutral SNP dataset (HO = 0.138, HE = 0.112, Na = 1.48; see Table S4).
Estimates of Ne for the different datasets (Tables 1, S4) were mostly consistent across all PCrit values used; when inconsistency among estimates was found, we retained the estimate obtained under the highest threshold (0.05). Reliable estimates could not be obtained for most ponds with the neutral SNP dataset, suggesting high population sizes in most ponds (except Pond 3, Table 1). Estimates based on the full SNP dataset were lower, with no reliable estimates for seven ponds and low values in most of the others (< 100, except for ponds 5 and 16, Table S4).
Population structure
Global FST as estimated with the neutral SNP dataset was 0.0233 (p-value = 0.001), and higher with the full SNP dataset (0.0052, p-value: 0.001), showing evidence of population structure in the study area. Focusing on the neutral SNP dataset, some pairwise FST values were significantly different from zero in comparisons involving certain ponds, especially ponds 3, 7, 11 and 14 (Tables S5–S6). Both STRUCTURE and DAPC analyses identified K = 2 as the optimal number of population units. At this clustering level, most demes had average assignment probabilities > 75% to a major genetic cluster widespread across the study area (Fig. S1). Similar results were obtained for the full SNP dataset (Fig. S2). DAPC results (Figs. S3–S6) were largely congruent with STRUCTURE, with individual assignment probabilities at K = 2 also showing a widespread genetic cluster (Cluster 1), with individuals from all populations, and a second cluster (Cluster 2) with, on average, higher assignment probabilities in the northernmost part of the study area (ponds 1–8) (Figs. S3–S4). Most ponds with lower assignment proportions to Cluster 2 are from the central (ponds 6 and 9) or southern (ponds 10–17) part of the study area, which is dominated by intensive agricultural landscapes. The scatterplot of DAPC results with ADEGENET showed little structure, with demes of ponds 1, 5, 6 and 17 being the most differentiated along the first two axes based on the neutral SNP dataset (Fig. S5).
The spatial clustering analysis with TESS provided finer resolution of genetic structure among ponds (Figs. 2, S7–9). Using the neutral SNP dataset, likelihood scores did not show either a clear minimum value or a plateau (Fig. S7), but 95% confidence intervals for runs with K > 6 were very broad, so results using K = 2 to K = 7 are presented (Figs. 2, S8). For K = 2, two genetic clusters are recovered showing some geographic structure, with a trend of decreasing average assignment probabilities to the minor genetic cluster towards the south of the study area. Results for other K values also suggest some differences between northern and southern population groups, with the intermediate Pond 14 showing some differentiation from its closest neighbors (Fig. 2). Results were similar with the full SNP dataset (Figure S9).
Landscape genomics
Based on Mantel test results, no evidence for IBD was found, neither with the neutral nor with the full SNP dataset and using FST (as FST/1 − FST) or DPS as measures of genetic distance, with no significant correlation between pairwise genetic and geographic distances (Figs. S10–S12). Regarding landscape genomics analyses, lower NDVI values in the study area were associated with low vegetation cover, as in traditional rainfed crops or grasslands, whereas high NDVI values corresponded to streams, which in the study area are mostly covered by riparian vegetation, and forests. The optimization of resistance surfaces using the neutral SNP dataset showed open areas associated with traditional agriculture, such as rainfed crops and grasslands, to be the land cover classes with higher conductance (Figs. 3, S13). Forest was the landscape feature showing higher resistance according to the optimized surface, with irrigated crops and artificial surfaces having intermediate conductance (Fig. S13). Both resistance optimization and model selection were similar in analyses with the full SNP dataset (Fig. S14–S15). Although all models other than the null were significant, the model including only isolation by resistance (IBR) had better explanatory power regarding patterns of connectivity among demes than models including IBD or the null model (Tables 2, S7).
NDVI values for the five coefficients used for optimization of resistance surfaces, and final conductance values after optimization with Genetic Algorithm with the goal of maximizing the connectivity using genetic data, with the neutral SNP dataset. The coefficient weights resulting from the optimization are shown for each NDVI coefficient
Discussion
Landscape genomics studies can take advantage of large SNP panels to answer questions on the relative roles of local demography and landscape features in shaping patterns of genetic diversity and population structure. Based on a set of 1390 neutral markers, we estimated low HO and HE values in all demes, raising concerns about the long-term viability of P. waltl populations in the study area. Low genetic diversity in these demes may be a consequence of both historical and recent processes. Our study area is located at the northern range limit for the species, with populations that are part of a genetic lineage that colonized the Iberian North Plateau relatively recently (in the last Inter-Glacial, about 120,000 years ago). This colonization process probably involved sequential bottleneck events, causing sharp reductions in several genetic diversity indices in comparison with populations located closer to historical refugia (Gutiérrez-Rodríguez et al. 2017b). Additional support for this hypothesis comes from the fact that preliminary screening of microsatellite markers in five ponds from our study area showed a large proportion of them to be monomorphic, although they were shown to be polymorphic in central Iberian populations. However, effective population sizes seem to be high, suggesting populations are not threatened by inbreeding depression and retain their evolutionary potential (Frankham et al. 2014), provided functional connectivity is maintained.
We found shallow but significant genetic structure among P. waltl demes in the study area, with no evidence of isolation by distance at the spatial scale considered, although landscape genomics analyses supported a role for geographic distance in shaping patterns of genetic differentiation (see below). Previous studies of the species showed low dispersal potential and strong genetic structure at small spatial scales (Gutiérrez-Rodríguez et al. 2017a, c; Fernández de Larrea et al. 2021; Reyes-Moya et al. 2022). Our results show less pronounced population differentiation than other amphibian studies conducted at similar or even smaller spatial scales (Oliver et al. 2017; Antunes et al. 2018; Sinai et al. 2019; Winiarski et al. 2019; Haugen et al. 2020; Van Buskirk and Van Rensburg 2020), even in environments with no apparent barriers to dispersal, including agricultural landscapes (Sotiropoulos et al. 2013; Hauguen et al. 2020; Unglaub et al. 2021).
Our landscape genomic analyses shed light on the main factors shaping genetic structure in the study area. Whereas IBR provided the best fit to the data, IBD (alone or in combination with IBR) also contributed to increase explanatory power, suggesting a role for geographic distance on differentiation. In addition, IBR results show higher landscape resistance at higher values of NDVI (Figs. 3, S15), supporting that open areas associated with traditional agricultural areas (including rainfed crops and grasslands) promote connectivity to a greater extent than forested areas. In addition to the flat topography and positive effect of open areas on gene flow in the study area, long distance movements of newts can be facilitated by some landscape features, especially wetlands and water courses (Da Fonte et al. 2019; Cayuela et al. 2020). During our field surveys, we observed individuals of P. waltl using streams and irrigation channels, suggesting that these linear water structures can act as corridors for amphibian dispersal, as reported in other studies (Emel and Storfer 2014; Haugen et al. 2020; Parsley et al. 2020). Information about dispersal in P. waltl is still incomplete, but it likely takes place mostly during their terrestrial stage and in juvenile phases, as in other urodeles (Perret et al. 2003; Roe and Grayson 2008). Therefore, flat and open areas, such as crops, may be preferred by the species during its dispersal phase. On the other hand, streams and areas with dense vegetation (high NDVI values) may be used as refuges during dry or cold periods but, based on our analyses, do not seem to be used as landscape corridors for dispersal in the study area.
An intensively managed zone, with mainly irrigated corn, is located in the study area between ponds 8 and 9, separating the traditionally managed rainfed area (ponds 1–7 and 10) and a heterogeneous cropland area comprising rainfed and irrigated crops (ponds 11–14). Different amphibian communities have been previously documented in these two areas (Albero et al. 2021), with less diversity and structure in the communities from the irrigated area, and P. waltl being less abundant than in the rainfed area. The irrigated area showed higher values of NDVI and lower conductance than traditional rainfed areas (Fig. 3), and TESS results show differences in genetic clustering assignments associated with this intensively managed area (for K = 2, see Fig. 2), suggesting it may act as a soft barrier restricting dispersal between the two major groups of ponds in the study area. These results are consistent with other studies reporting higher genetic differentiation in amphibian populations in intensive agricultural landscapes compared to natural habitats (Lenhardt et al. 2017; Gauffre et al. 2022; but see Youngquist et al. 2017). Nevertheless, given the long generation time of P. waltl (Cayuela et al. 2022), the effects of the recent shift to irrigation in this area on patterns of genetic structure may not be fully appreciable yet.
The shallow genetic structure documented in the present study contrasts with the results of Gutiérrez-Rodríguez et al. (2017c), who assessed the genetic structure of P. waltl populations in central Spain at a similar geographic scale, finding strong differentiation over short distances (5–10 km). Apart from differences in the historical demography of their sampled populations, which are closer to historical glacial refugia and thus are more genetically diverse than those in our study, and in the type of markers used (microsatellites vs. SNPs), landscape features differ substantially between both studies and have probably contributed to shape patterns of connectivity among demes differently, as shown by our landscape genomics analyses. For instance, the dominant land cover class in Gutiérrez-Rodríguez et al. (2017c) is “dehesa” woodland and scrub, with a minor proportion of agricultural land cover classes, which dominate our study area. More importantly, their study was conducted in the foothills of Sierra de Guadarrama, a hilly area, with slope and altitude being recovered as two of the major factors restricting gene flow, in agreement with studies in other amphibian species (Sánchez-Montes et al. 2018; Cayuela et al. 2020). In contrast, our study area is part of a flat plateau with no significant topographic features, which may favor gene flow. Gutiérrez-Rodríguez et al. (2017c) also found a positive role of the spatial heterogeneity of vegetation amount (NDVI st.-dev), rather than of vegetation cover itself. These contrasting results highlight the fact that the relationship between landscape features and gene flow is often complex and can vary throughout species’ ranges, depending on the availability and spatial configuration of different types of terrestrial habitat patches as well as on historical factors. Another important feature potentially contributing to differences between studies in inferred patterns of connectivity among demes is the higher pond density in our study area (~ 1 pond/10 km2, see Fig. 1). Most of these ponds are suitable breeding habitats for the target species, and most of them are surrounded by agricultural plots instead of forest, which can partly explain why agricultural areas showed lower resistance to gene flow. In this context, climate change projections for the Mediterranean basin predict a decrease in precipitation and an increasing incidence of heat waves, which will likely result in more frequent and severe drought periods (Giorgi and Lionello 2008) and a concomitant decrease in pond hydroperiod and density at the landscape scale. Understanding the role of different factors in promoting/restricting connectivity among demes is therefore fundamental to design and implement conservation plans for Mediterranean amphibian communities in the face of climate change.
Few studies have compared patterns of genetic structure of passive vs. active dispersing organisms in aquatic metacommunities. Using presence/absence data, ecological studies have found a stronger spatial effect, linked to dispersal limitations, for both passive dispersers (macrophytes, zooplankton) and large-bodied active dispersers (amphibians), in contrast to flying insects, which show no dispersal limitation (Van De Meutter et al. 2007; De Bie et al. 2012). Previous studies on a passive-dispersal aquatic plant species (Myriophyllum alterniflorum) in the same ponds sampled in this study revealed a marked genetic discontinuity between the pond groups of Payuelos and Oteros, with low levels of gene flow and a strong IBD pattern (García-Girón et al. 2019), in agreement with this expectation. Our results show overall higher connectivity in P. waltl than in Myriophyllum, highlighting that under favorable conditions, amphibian populations can remain functionally connected, even in humanized landscapes.
In conclusion, our study highlights the important role of landscape features, such as open areas resulting from traditional rainfed agriculture, in promoting functional connectivity of amphibian populations in Mediterranean agrosystems. Temporary ponds are unique ecosystems that are increasingly threatened by urban and agricultural development and the introduction of alien species (Beja and Alcazar 2003). Conservation guidelines and policies such as the Common Agricultural Policy or the Water Framework Directive (both from the European Union) should adopt a functional network strategy, protecting clusters of inter-connected temporary ponds throughout the agricultural matrix and landscape features that favor connectivity, such as traditional rainfed crops, to ensure the conservation of the diverse amphibian communities of Mediterranean ecosystems.
References
Albero L, Martínez-Solano Í, Arias A, Lizana M, Bécares E (2021) Amphibian metacommunity responses to agricultural intensification in a Mediterranean landscape. Land 10:924
Antunes B, Lourenço A, Caeiro-Dias G, Dinis M, Gonçalves H, Martínez-Solano Í, Velo-Antón G (2018) Combining phylogeography and landscape genetics to infer the evolutionary history of a short-range Mediterranean relict, Salamandra salamandra longirostris. Conserv Genet 19(6):1411–1424
Beja P, Alcazar R (2003) Conservation of Mediterranean temporary ponds under agricultural intensification: an evaluation using amphibians. Biol Conserv 114(3):317–326
Beja P, Bosch J, Tejedo M, Edgar P, Donaire-Barroso D, Lizana M, Martínez-Solano I, Salvador A, García-París M, Recuero Gil E, Slimani T, El Mouden EH, Geniez P, Slimani T (2009) Pleurodeles waltl. In: The IUCN Red List of Threatened Species 2009. e.T59463A11926338
Beklioglu M, Romo S, Kagalou I, Quintana X, Becares E (2007) State of the art in the functioning of shallow Mediterranean lakes: workshop conclusions. Hydrobiologia 584:317–326
Benton TG, Vickery JA, Wilson JD (2003) Farmland biodiversity: is habitat heterogeneity the key? Trends Ecol Evol 18(4):182–188
Bolle HJ (2003) Mediterranean climate. Variability and trends. Springer, Berlin
Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA (2013) Stacks: an analysis tool set for population genomics. Mol Ecol 22(11):3124–3140
Caye K, Deist TM, Martins H, Michel O, François O (2016) TESS3: fast inference of spatial population structure and genome scans for selection. Mol Ecol Resour 16(2):540–548
Cayuela H, Valenzuela-Sánchez A, Teulier L, Martínez-Solano Í, Léna JP, Merilä J, Muths E, Shine R, Quay L, Denoël M, Clobert J, Schmidt BR (2020) Determinants and consequences of dispersal in vertebrates with complex life cycles: a review of pond-breeding amphibians. Q Rev Biol 95:1–36
Cayuela H, Lemaitre J-F, Léna J-P, Ronget V, Martínez-Solano I, Muths E, Pilliod DS, Schmidt BR, Sánchez-Montes G, Gutiérrez-Rodríguez J, Pyke G, Grossenbacher K, Lenzi O, Bosch J, Beard KH, Woolbright LL, Lambert BA, Green DM, Garwood JM, Fisher RN, Matthews K, Dudgeon D, Lau A, Speybroeck J, Homan R, Jehle R, Baskale E, Mori E, Arntzen JW, Joly P, Stiles RM, Lannoo MJ, Maerz JC, Lowe WH, Valenzuela-Sánchez A, Christiansen DG, Angelini C, Thirion J-M, Merilä J, Colli GR, Vasconcellos MM, Boas TCV, Arantes IC, Levionnois P, Reinke BA, Vieira C, Marais GAB, Gaillard J-M, Miller DAW (2022) Sex-related differences in aging rate are associated with sex chromosome system in amphibians. Evolution 76:346–356
Chang CC, Chow CC, Tellier LCAM, Vattikuti S, Purcell SM, Lee JJ (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4(1):1–16
Córdova-Lepe F, Del Valle R, Ramos-Jiliberto R (2018) The process of connectivity loss during habitat fragmentation and their consequences on population dynamics. Ecol Modell 376:68–75
Da Fonte LFM, Mayer M, Lötters S (2019) Long-distance dispersal in amphibians. Front Biogeogr 11(4):1–14
De Bie T, De Meester L, Brendonck L, Martens K, Goddeeris B, Ercken D, Hampel H, Denys L, Vanhecke L, Van der Gucht K, Van Wichelen J, Vyverman W, Declerck SAJ (2012) Body size and dispersal mode as key traits determining metacommunity structure of aquatic organisms. Ecol Lett 15:740–747
Do C, Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR (2014) NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour 14(1):209–214
Earl DA, Von Holdt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4(2):359–361
Elewa A, Wang H, Talavera-López C, Joven A, Brito G, Kumar A, Hameed LS, Penrad-Mobayed M, Yao Z, Zamani N, Abbas Y, Abdullayev I, Sandberg R, Grabherr M, Andersson B, Simon A (2017) Reading and editing the Pleurodeles waltl genome reveals novel features of tetrapod regeneration. Nat Commun 8:1–9
Emel SL, Storfer A (2014) Landscape genetics and genetic structure of the southern torrent salamander Rhyacotriton variegatus. Conserv Genet 16(1):209–221
Estrada-Peña A, Estrada-Sánchez A, de la Fuente J (2014) A global set of Fourier-transformed remotely sensed covariates for the description of abiotic niche in epidemiological studies of tick vector species. Parasite Vectors 7(1):1–14
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14(8):2611–2620
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10(3):564–567
Fahrig L, Baudry J, Brotons L, Burel FG, Crist TO, Fuller RJ, Sirami C, Siriwardena GM, Martin JL (2011) Functional landscape heterogeneity and animal biodiversity in agricultural landscapes. Ecol Lett 14:101–112
Fardila D, Kelly LT, Moore JL, McCarthy MA (2017) A systematic review reveals changes in where and how we have studied habitat loss and fragmentation over 20 years. Biol Conserv 212:130–138
Fernández Aláez M, Fernández Aláez C, Rodríguez S, Bécares E (1999) Evaluation of the state of conservation of shallow lakes in the province of Leon (Northwest Spain) using botanical criteria. Limnetica 17:107–117
Fernández de Larrea I, Sánchez-Montes G, Gutiérrez-Rodríguez J, Martínez-Solano Í (2021) Reconciling direct and indirect estimates of functional connectivity in a Mediterranean pond-breeding amphibian. Conserv Genet 22:455–463
Ferreira M, Beja P (2013) Mediterranean amphibians and the loss of temporary ponds: are there alternative breeding habitats? Biol Conserv 165:179–186
Ficetola GF, De Bernardi F (2004) Amphibians in a human-dominated landscape: the community structure is related to habitat features and isolation. Biol Conserv 119(2):219–230
Foley JA, DeFries R, Asner GP, Barford C, Bonan G, Carpenter SR, Chapin FS, Coe MT, Daily GC, Gibbs HK, Helkowski JH, Holloway T, Howard EA, Kucharik CJ, Monfreda C, Patz JA, Prentice IC, Ramankutty N, Snyder PK (2005) Global consequences of land use. Science 309:570
Foll M, Gaggiotti O (2008) A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180:977–993
Frankham R, Bradshaw CJ, Brook BW (2014) Genetics in conservation management: revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biol Conserv 170:56–63
García-Girón J, García P, Fernández-Aláez M, Bécares E, Fernández-Aláez C (2019) Bridging population genetics and the metacommunity perspective to unravel the biogeographic processes shaping genetic differentiation of Myriophyllum alterniflorum DC. Sci Rep. https://doi.org/10.1038/s41598-019-54725-7
Gauffre B, Boissinot A, Quiquempois V, Leblois R, Grillet P, Morin S, Picard D, Ribout C, Lourdais O (2022) Agricultural intensification alters marbled newt genetic diversity and gene flow through density and dispersal reduction. Mol Ecol 31:119–133
Giorgi F, Lionello P (2008) Climate change projections for the Mediterranean region. Glob Planet Change 63(2–3):90–104
Gray MJ, Smith LM, Leyva RI (2004) Influence of agricultural landscape structure on a Southern High Plains, USA, amphibian assemblage. Landsc Ecol 19(7):719–729
Gutiérrez-Rodríguez J, Sánchez-Montes G, Martínez-Solano I (2017a) Effective to census population size ratios in two Near Threatened Mediterranean amphibians: Pleurodeles waltl and Pelobates cultripes. Conserv Genet 18(5):1201–1211
Gutiérrez-Rodríguez J, Barbosa AM, Martínez-Solano Í (2017b) Integrative inference of population history in the Ibero-Maghrebian endemic Pleurodeles waltl (Salamandridae). Mol Phylogenet Evol 112:122–137
Gutiérrez-Rodríguez J, Gonçalves J, Civantos E, Martínez-Solano I (2017c) Comparative landscape genetics of pond-breeding amphibians in Mediterranean temporal wetlands: the positive role of structural heterogeneity in promoting gene flow. Mol Ecol 26(20):5407–5420
Haddad NM, Brudvig LA, Clobert J, Davies KF, Gonzalez A, Holt RD, Lovejoy TE, Sexton JO, Austin MP, Collins CD, Cook WM, Damschen EI, Ewers RM, Foster BL, Jenkins CN, King AJ, Laurance WF, Levey DJ, Margules CR, Townshend JR (2015) Habitat fragmentation and its lasting impact on earth’s ecosystems. Sci Adv 1(2):1–10
Halverson MA, Skelly DK, Caccone A (2006) Inbreeding linked to amphibian survival in the wild but not in the laboratory. J Hered 97:499–507
Haugen H, Linløkken A, Østbye K, Heggenes J (2020) Landscape genetics of northern crested newt Triturus cristatus populations in a contrasting natural and human-impacted boreal forest. Conserv Genet 21(3):515–530
Jombart T (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24(11):1403–1405
Jombart T (2012) A tutorial for Discriminant Analysis of Principal Components (DAPC) using adegenet. MRC Centre for Outbreak Analysis and Modelling, Imperial College, London
Keenan K, McGinnity P, Cross TF, Crozier WW, Prodöhl PA (2013) diveRsity: an R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol Evol 4(8):782–788
Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15(5):1179–1191
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10(3):R25
Lenhardt PP, Brühl CA, Leeb C, Theissinger K (2017) Amphibian population genetics in agricultural landscapes: does viniculture drive the population structuring of the European common frog (Rana temporaria)? PeerJ 5:e3520
Luu K, Bazin E, Blum MGB (2017) Pcadapt: an R package to perform genome scans for selection based on principal component analysis. Mol Ecol Resour 17:67–77
Marchi C, Andersen LW, Damgaard C, Olsen K, Jensen TS, Loeschcke V (2013) Gene flow and population structure of a common agricultural wild species (Microtus agrestis) under different land management regimes. Heredity 111(6):486–494
Maroso F, De Gracia CP, Iglesias D, Cao A, Díaz S, Villalba A, Vera M, Martínez P (2019) A useful snp panel to distinguish two cockle species, Cerastoderma edule and C. glaucum, co-occurring in some European beds, and their putative hybrids. Genes 10:760
McCartney-Melstad E, Vu JK, Shaffer HB (2018) Genomic data recover previously undetectable fragmentation effects in an endangered amphibian. Mol Ecol 27:4430–4443
Newbold T, Hudson LN, Hill SLL, Contu S, Lysenko I, Senior RA, Börger L, Bennett DJ, Choimes A, Collen B, Day J, De Palma A, Díaz S, Echeverria-Londoño S, Edgar MJ, Feldman A, Garon M, Harrison MLK, Alhusseini T, Ingram DJ, Itescu Y, Kattge J, Kemp V, Kirkpatrick L, Kleyer M, Correia DLP, Martin CD, Meiri S, Novosolov M, Pan Y, Phillips HRP, Purves DW, Robinson A, Simpson J, Tuck SL, Weiher E, White HJ, Ewers RM, MacE GM, Scharlemann JPW, Purvis A (2015) Global effects of land use on local terrestrial biodiversity. Nature 520:45–50
Oksanen J, Guillaume BF, Kindt R, Legendre P, Minchin P, O’Hara R (2017) vegan: Community ecology package. R package version 2.3-3. https://cran.r-project.org/web/packages/vegan/index.html
Oliver V, Catherine SG, Jinzhong I (2017) Syntopic frogs reveal different patterns of interaction with the landscape: a comparative landscape genetic study of Pelophylax nigromaculatus and Fejervarya limnocharis from central China. Ecol Evol 7:9294–9306
Parsley MB, Torres ML, Banerjee SM, Tobias ZJC, Goldberg CS, Murphy MA, Mims MC (2020) Multiple lines of genetic inquiry reveal effects of local and landscape factors on an amphibian metapopulation. Landsc Ecol 35(2):319–335
Pembleton LW, Cogan NOI, Forster JW (2013) StAMPP: an R package for calculation of genetic differentiation and structure of mixed-ploidy level populations. Mol Ecol Resour 13:946–952
Perret N, Pradel R, Miaud C, Grolet O, Joly P (2003) Transience, dispersal and survival rates in newt patchy populations. J Anim Ecol 72:567–575
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Prive F, Luu K, Vilhjalmsson BJ, Blum MGB (2020) Performing highly efficient genome scans for local adaptation with R Package pcadapt Version 4. Mol Biol Evol 37:2153–2154
Reyes-Moya I, Sánchez-Montes G, Martínez-Solano I (2022) Integrating dispersal, breeding and abundance data with graph theory for the characterization and management of functional connectivity in amphibian pondscapes. Landsc Ecol 37:3159–3177
Rödin-Mörch P, Palejowski H, Cortazar-Chinarro M, Kärvemo S, Richter-Boix A, Höglund J, Laurila A (2021) Small-scale population divergence is driven by local larval environment in a temperate amphibian. Heredity 126(2):279–292
Roe AW, Grayson KL (2008) Terrestrial movements and habitat use of juvenile and emigrating adult eastern red- spotted newts, Notophthaltnus viridescens. Soc Study Amphib Reptiles 42(1):22–30
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219–1228
Rousset F (2008) GENEPOP’007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour 8(1):3–106
Salvador A (2015) Gallipato - Pleurodeles waltl. In: Salvador A, Martínez-Solano I (eds) Enciclopedia Virtual de los Vertebrados Españoles. Museo Nacional de Ciencias Naturales, Madrid
Sánchez-Montes G, Wang J, Ariño AH, Martínez-Solano Í (2018) Mountains as barriers to gene flow in amphibians: quantifying the differential effect of a major mountain ridge on the genetic structure of four sympatric species with different life history traits. J Biogeogr 45(2):318–331
Sinai I, Segev O, Weil G, Oron T, Merilä J, Templeton AR, Blaustein L, Greenbaum G, Blank L (2019) The role of landscape and history on the genetic structure of peripheral populations of the Near Eastern fire salamander, Salamandra infraimmaculata Northern Israel. Conserv Genet 20(4):875–889
Smith AL, Gardner MG, Fenner AL, Bull CM (2009) Restricted gene flow in the endangered pygmy bluetongue lizard (Tiliqua adelaidensis) in a fragmented agricultural landscape. Wildl Res 36(6):466–478
Sotiropoulos K, Eleftherakos K, Tsaparis D, Kasapidis P, Giokas S, Legakis A, Kotoulas G (2013) Fine scale spatial genetic structure of two syntopic newts across a network of ponds: implications for conservation. Conserv Genet 14:385–400
Spurgin LG, Gage MJG (2019) Conservation: the costs of inbreeding and of being inbred. Curr Biol 29(16):R796–R798
Storfer A, Murphy MA, Spear SF, Holderegger R, Waits LP (2010) Landscape genetics: where are we now? Mol Ecol 19(17):3496–3514
Taylor PD (2006) Landscape connectivity: a return to the basics. In: Sanjayan MA (ed) Connectivity conservation. Cambridge University Press, Cambridge, pp 29–43
Unglaub B, Cayuela H, Schmidt BR, Preißler K, Glos J, Steinfartz S (2021) Context-dependent dispersal determines relatedness and genetic structure in a patchy amphibian population. Mol Ecol 30(20):5009–5028
Van Buskirk J, Jansen van Rensburg A (2020) Relative importance of isolation-by-environment and other determinants of gene flow in an alpine amphibian. Evolution 74(5):962–978
Van De Meutter F, De Meester L, Stoks R (2007) Metacommunity structure of pond macroinvertebrates: effects of dispersal mode and generation time. Ecology 88(7):1687–1695
Velo-Antón G, Lourenço A, Galán P, Nicieza A, Tarroso P (2021) Landscape resistance constrains hybridization across contact zones in a reproductively and morphologically polymorphic salamander. Sci Rep 11(1):1–16
Vos CC, Goedhart PW, Lammertsma DR, Spitzen-Van Der Sluijs AM (2007) Matrix permeability of agricultural landscapes: an analysis of movements of the common frog (Rana temporaria). Herpetol J 17(3):174–182
Wang S, Meyer E, Mckay JK, Matz MV (2012) 2b-RAD: a simple and flexible method for genome-wide genotyping. Nat Methods 9(8):808–810
Winiarski KJ, Mcgarigal K, Peterman WE, Whiteley AR (2019) Multiscale resistant kernel surfaces derived from inferred gene flow : an application with vernal pool breeding salamanders. Mol Ecol Resour 00:1–17
Wright S (1931) Evolution in Mendelian populations. Genetics 16:97–159
Youngquist MB, Inoue K, Berg DJ, Boone MD (2017) Effects of land use on population presence and genetic structure of an amphibian in an agricultural landscape. Landsc Ecol 32(1):147–162
Acknowledgements
We thank Omar Rodríguez, Adrián Llamazares, María Borrego, Víctor Ezquerra, Pablo Oviedo, Alejandro Cachorro, Rubén de Prado and especially Ana Arias and Alberto Benito for their help during fieldwork and Lucía Insua for her technical support. Andrés Blanco, Adrián Casanova, Steve Mussmann, Gregorio Sánchez Montes, Ángel Gálvez and Ismael Reyes provided helpful feedback on genetic analyses and interpretation of results, and three anonymous reviewers contributed constructive comments that considerably improved the manuscript. Erik Wild improved the final text. We also thank the SCAYLE Foundation for their support in bioinformatic analysis.
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature. This research was supported by the Spanish Ministerio de Economía, Industria y Competitividad, project Metaponds (ref: CGL2017-84176-R); by the Fundación Biodiversidad, Ministerio para la Transición ecológica y el Reto Demográfico (project BT-2019) and by the University of León (Convocatoria 2022 de Ayudas a Proyectos de Investigación competitivos). L. Albero is funded by a PhD grant from Universidad de León (ULE).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Albero, L., Martínez-Solano, Í., Hermida, M. et al. Open areas associated with traditional agriculture promote functional connectivity among amphibian demes in Mediterranean agrosystems. Landsc Ecol 38, 3045–3059 (2023). https://doi.org/10.1007/s10980-023-01725-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10980-023-01725-8