Abstract
Context
Mediterranean wetland ecosystems are in continuous decline due to human pressure. Amphibians are key elements of biotic communities of Mediterranean temporary ponds and streams, and their persistence depends on the availability and inter-connectivity of breeding sites.
Objectives
We investigated the role of different factors potentially driving functional connectivity patterns in two amphibian species at the landscape and local scales. We focused on two Mediterranean endemic pond-breeding amphibians inhabiting semi-arid landscapes of central Spain, the common parsley frog (Pelodytes punctatus) and the common midwife toad (Alytes obstetricans).
Methods
We genotyped 336 individuals of P. punctatus and 318 of A. obstetricans from 17 and 16 breeding populations at 10 and 17 microsatellite loci, respectively. We used remotely sensed vegetation/moisture indices and land use/cover data to derive optimized resistance surfaces and test their association with estimates of gene flow and migration rates across populations.
Results
We found evidence for higher population connectivity in common midwife toads than in common parsley frogs, with a strong effect of water availability in patterns of population connectivity of both species. However, the two species differ in the role of landscape features on population connectivity, with the distance and spatial distribution of artificial land-use types positively influencing connectivity in A. obstetricans and meadows/pastureland favouring P. punctatus. This is in line with reported breeding site preferences for the two species, with A. obstetricans successfully breeding in artificial water bodies that P. punctatus generally avoid.
Conclusions
This study highlights the importance of assessing species–habitat relationships shaping connectivity when developing and implementing conservation and management actions to benefit fragmented amphibian populations in the Mediterranean region. Our results show that amphibian species respond differently, even contrastingly to landscape features and thus require alternative, complementary strategies to improve population connectivity and ensure long-term viability.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Mediterranean region is characterized by the presence of ecologically diverse types of wetlands (Pearce and Crivelli 1994; Médail and Quézel 1999). These ecosystems are abundant in southern Europe and North Africa and are listed as EU Priority Habitat (Natura 2000 code 3170, 92/43/CEE, 21 May 1992), but are in continuous decline due to the impact of human activities (Green et al. 2002; Bouahim et al. 2010). This decline is expected to be exacerbated by climate change, which will have a much greater impact on smaller, temporary water bodies (Parmesan 2006; Rhazi et al. 2012). Ephemeral wetlands in endorheic depressions are characterized by high seasonality and fluctuating water levels linked to extremely variable precipitation patterns across seasons and years (Vanschoenwinkel et al. 2009; Sahuquillo et al. 2012). These habitats host diverse flora and fauna communities (Williams 2006), including many rare and threatened species (Keddy 2010), which are adapted to living under the stress associated with these extreme environmental conditions (Grillas et al. 2004).
Amphibians are key elements in the biotic communities of Mediterranean temporary ponds and streams (Gómez-Rodríguez et al. 2009). At the local scale, their breeding success and population dynamics directly depend on the availability of water bodies and their hydroperiod (Newman 1992; Jakob et al. 2003; Gómez-Rodríguez et al. 2010a, b, c; Cayuela et al. 2012). At the landscape scale, their persistence depends on the availability and connectivity among breeding sites, which is a function of the dispersal ability of the different species and the resistance of the terrestrial habitat matrix to movement.
The loss of Mediterranean temporary ponds and streams has had a significant impact on biodiversity (Cuttelod et al. 2009), and is one of the major threats causing amphibian declines in the region (Stuart et al. 2004, 2008). In areas where the availability of natural breeding sites is low because of changes in land use, aridization and desertification, like central Iberia, some amphibian species can take advantage of artificial constructions to hold water for irrigation, and breed successfully (Caballero-Díaz et al. 2020). However, little is known about patterns of connectivity in amphibian populations in these regions. Identifying factors shaping functional connectivity patterns (i.e., gene flow) in these areas is critical to guide management efforts to conserve amphibian populations and their associated biotic communities in view of the increasing fragmentation of their populations (Martínez-Solano 2006).
Landscape genetic tools allow linking genetic data with different potential response variables and explicitly quantify the effects of landscape composition, configuration and matrix quality on spatial patterns of genetic variation and gene flow (Storfer et al. 2007). Assessing comparative patterns of regional gene flow in co-distributed species can help develop cost-effective management practices with measures favouring a broader set of taxa (Gutiérrez-Rodríguez et al. 2017). Here, we focused on two Mediterranean endemic pond-breeding amphibians, the common parsley frog (Pelodytidae: Pelodytes punctatus), and the common midwife toad (Alytidae: Alytes obstetricans). These species have broadly overlapping distributions but rarely share breeding sites. Pelodytes punctatus prefers shallow temporary ponds and streams, including those formed in flooded quarries and cultivated areas (Denoël et al. 2009; Caballero-Díaz et al. 2020), whereas A. obstetricans preferentially uses stagnant permanent ponds and pools, including water tanks and other artificial sites (Bosch et al. 2009; Caballero-Díaz et al. 2020). Both species are listed as Least Concern (LC) by the IUCN, but their populations are in decline globally and locally in the study area due to a combination of factors, including habitat loss and the negative effect of alien species and emerging infectious diseases (Martínez-Solano 2006; Bosch et al. 2009; Denoël et al. 2009; Caballero-Díaz et al. 2020).
We investigated the role of different factors potentially driving functional connectivity patterns in both species at the landscape and local scales. We genotyped a large sample of individuals of both species in a geographically comprehensive dataset and used remotely sensed vegetation/moisture indices and discrete land use/cover categories to derive optimized resistance surfaces to test their association with estimates of gene flow and migration rates across populations. We expected differences in the role of landscape features on connectivity patterns across species because they differ in their breeding site preferences, with P. punctatus usually occupying shallow temporary ponds and A. obstetricans generally using permanent water bodies, including artificial sites like water tanks used for irrigation. Finally, we discuss potential applications of our results to the design of management actions that favour the persistence of viable population networks at the regional scale.
Material and methods
Study area and sampling
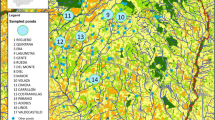
The study area is located in the centre of Iberian Peninsula, bounded by − 3.784º to 3.024º longitude and 39.813º to 40.571º latitude (Fig. 1). Elevation ranges from 470 m up to 940 m.a.s.l., and the Jarama, Tajuña and Tagus rivers shape a complex orography, characterized by paramos with loam, gypsum and limestone in calcareous soils. Paramos are dominated by olive groves, vineyards and cereal crops, with corn in irrigated areas along river valleys. Typical Mesomediterranean vegetation occupies a reduced extension with the presence of pine forests, holm oaks (Quercus ilex), gallery forests and extensions of salt cedar (Tamarix sp.), with scrublands characterized by the presence of kermes oaks (Quercus coccifera) (Izco 1984). The two amphibian species selected for this study are broadly but discontinuously distributed in the region (Caballero-Díaz et al. 2020). We sampled 336 individuals of P. punctatus and 318 of A. obstetricans at 17 and 16 localities, respectively (Fig. 1, Table 1). Because both species are philopatric and tend to congregate in the vicinities of breeding sites, the sampling was focused on breeding populations, which are clearly delimited in the study area by the availability of water bodies and are largely isolated from each other by large extensions of unsuitable terrestrial habitat. Tissue samples were taken from tail clips of tadpoles of both species, which were collected using dip nets. In order to minimize the probability of sampling siblings, we captured tadpoles of different sizes (potentially representing different cohorts) and in different sections of the water bodies.
Sampling locations for the two study species: P. punctatus and A. obstetricans. Population codes as in Table 1. A Land use map of the study area. B Topographic map of the study area. The inset in B shows the location of the study area in central Spain
Average geographic distances across populations were 4.97 ± 3.25 km in Alytes and 7.12 ± 3.99 km in Pelodytes (Fig. 1; Supplementary material Tables S1 and S2). Tissue samples for molecular analyses were preserved in 100% ethanol prior to DNA extraction.
Genetic analyses
Genomic DNA was extracted using NucleoSpin Tissue-Kits (Macherey–Nagel). A total of 10 previously characterized microsatellite markers for P. punctatus were amplified following published PCR conditions (Jourdan-Pineau et al. 2009; van de Vliet et al. 2009). These loci were grouped in three multiplex reactions (multiplex 1: PPU5, Ppu10, Ppu8, Ppu11; multiplex 2: PPU16, Ppu7, Ppu5; multiplex 3: Ppu14, Ppu15, Ppu9). For A. obstetricans, 17 previously published loci were grouped in five multiplex reactions (Maia‐Carvalho et al. 2014), and PCR conditions and genotype calling followed Maia‐Carvalho et al. (2014).
We tested for the presence of null alleles, stuttering and large allele dropout in microsatellite markers using MICROCHECKER v2.2.3 (van Oosterhout et al. 2004). Deviations from Hardy–Weinberg equilibrium (HWE) and evidence of linkage disequilibrium (LD) were estimated with the software GENEPOP (Raymond and Rousset 1995; Rousset 2008), applying the sequential Bonferroni correction (Rice 1989) to adjust significance values for multiple tests. We calculated different estimates of genetic diversity for each population using GENALEX v6.5b5 (Peakall and Smouse 2012), including the number of alleles (NA), observed (HO) and expected heterozygosity (HE). We also calculated the inbreeding coefficient (FIS) because it is an indirect measure of philopatry (more philopatric species will, in principle, have higher FIS values).
We applied three approaches to infer functional connectivity (= gene flow) from genetic data. First, we used BAPS v6 (Corander et al. 2008; Cheng et al. 2013) to characterize regional population genetic structure for both species. BAPS treats allele frequencies of molecular markers and the number of genetic clusters as random variables and uses Bayesian inference to characterize population genetic structure. We ran spatial genetic mixture analyses with ten independent runs and a maximum number of groups equal to the number of sampled localities. We compared genetic clusters resulting from each replicate run based on their likelihood score and identified the optimal clustering level based on a stochastic optimization algorithm (Corander et al. 2008).
Second, we calculated three indices of genetic differentiation between populations, FST (Weir and Cockerham 1984), G’’ST (Meirmans and Hedrick 2011) and DST (Jost 2008) using Genepop, GENODIVE v2.0b23 (Meirmans and van Tienderen 2004) and SMOGD v1.2.5 (Crawford 2010), respectively, to characterize patterns of genetic differentiation between populations of the two species. Measures of genetic differentiation provide indirect estimates of gene flow and may also reflect other processes, like genetic drift, but provide information about population structure at the landscape scale in philopatric organisms like amphibians. The three indices provide complementary information about population structure in different demographic and mutational scenarios (Jost et al. 2018).
Finally, we estimated recent migration rates between populations using BAYESASS v3.0 (Wilson and Rannala 2003). This program infers the proportion of recent migrants among populations from multilocus genotype data. We ran three different replicate analyses for each species with 50,000,000 iterations, with a burn-in period of 2,000,000 and sampling pairwise migration rate estimates each 2,000 iterations. We assessed convergence of results across runs using software TRACER (Rambaut et al. 2018) and used those with the best likelihood in subsequent analyses.
Input data used for resistance surfaces and pre-processing
Connectivity is influenced by several factors operating at distinct spatial and temporal scales (Auffret et al. 2015; Gonçalves et al. 2016). To address this, we identified the most relevant factors impacting landscape connectivity and gene flow based on previous literature (e.g., Pérez-Espona et al. 2008; Gutiérrez-Rodríguez et al. 2017; Lourenço et al. 2019). This process allowed us to identify several landscape dimensions, including topography/geomorphology, vegetation amount and heterogeneity, water/moisture availability, linear elements (either natural, like rivers, or artificial, like roads or motorways) and land use/cover (LUC). Given the importance of the latter for explaining connectivity in complex landscape mosaics with marked urban/peri-urban gradients, we used fine-scale LUC data from the Spanish Land Cover Information System for 2015—reclassified for simplicity—(Supplementary Material Table S3; URL: https://www.siose.es/) to compute several types of resistance surfaces based on: (i) the distance to patches of specific classes (e.g. urban areas, water surfaces), (ii) binary surfaces denoting the presence or absence of a certain LUC class, and (iii) the percentage cover by LUC category. All resistance surfaces were calculated at a spatial resolution of 100 m (i.e., pixel size), considered adequate to represent landscape/environmental patterns and with enough detail to evaluate connectivity and gene flow of target species within reasonable computing times.
For topographic variables, we employed a 25 m Digital Elevation Model from the Spanish Centre of Geographic Information (URL: http://centrodedescargas.cnig.es) to derive an elevation resistance surface. This data was also used to calculate several indices, including the slope, the Topographic Ruggedness Index as a descriptor of topographic/elevation complexity (TRI; at two aggregation kernels, k = 3 × 3 and k = 9 × 9 pixels), and the Topographic Wetness Index (TWI) describing the tendency of a cell to accumulate water (Quinn et al. 1995).
We also calculated river density (m/Km2) using data on the hydrographic network from the European Catchment Database (URL: https://www.eea.europa.eu/data-and-maps/data/european-river-catchments-1) for (i) all rivers in a 1000 m buffer, (ii) all rivers in a 2500 m buffer and, (iii) small rivers with Strahler class 1 or 2 in a 2500 m buffer.
Freely accessible Open Street Maps (OSM) data on the road network (URL: https://download.geofabrik.de/europe/spain.html) were employed to calculate: (i) road density, (ii) distance to roads and (iii) binary layers signalling the presence/absence of roads in each 100 m cell. To assess differences in connectivity response by road class and traffic volume, we approximated this based on OSM metadata, more specifically, the type of road. Motorways correspond to ‘high-traffic’ (typically with two or more lanes in each way) while other roads to ‘medium-traffic’ roads (typically with one lane per direction).
From Landsat-8 satellite data (USGS/NASA; URL: https://earthexplorer.usgs.gov/) with a spatial resolution of 30 m (16-day temporal resolution), we calculated resistance surfaces related to vegetation/greenness using the Normalized Difference Vegetation Index (NDVI; combining the red and near-infrared spectral bands), and related to water availability in soil/vegetation based on the Normalized Difference Water/Moisture Index (NDWI; combining spectral bands in the near-infrared and shortwave-infrared). Both NDVI and NDWI vary from − 1 to 1, with lower values indicating, respectively, low vegetation cover (e.g., non-vegetated/artificial surfaces) or low water availability, and values closer to one the opposite (e.g. densely vegetated areas). Image data, covering the months of in-situ sampling, were aggregated to 100 m, thus using the same spatial resolution of other resistance surfaces through the average (capturing the amount of vegetation cover or water in soil/vegetation) and the standard deviation (related to the spatial heterogeneity of the same aspects). A total of 54 different resistance surfaces were tested, optimized and ranked using a modelling approach (the complete list of resistance surfaces is detailed in Table 2).
Statistical modelling and resistance surface optimization
To assess the relative support of each resistance surface (or variable) to explain differences in genetic structure between sampled populations, we used the ResistanceGA package (Peterman et al. 2014; Peterman 2018) implemented in R v4.0 (R Core Team 2020). This R package implements an optimization approach based on genetic algorithms that selects the best numerical transformation type for a given resistance surface while also optimizing the equation parameters used in those transformations. ResistanceGA package starts with ‘raw’/untransformed surfaces (which display different landscape features such as road density or the % cover of meadows) and ends up with actual resistance surfaces which are tailored to the response of each feature and species. Optimization allows bypassing the subjective assignment of resistance surface values based on expert opinion or by modelling individual movement patterns (Peterman 2018). The resulting optimized surface is then applied to iteratively calculate the cost-based distance between sampled locations using least-cost path analyses through the gdistance package (van Etten 2017). ResistanceGA then applies a linear mixed effects model with the maximum likelihood population effects (MLPE) parameterization (Clarke et al. 2002) to relate the genetic and the cost-based distance matrices. Model inference and ranking of each resistance surface is based on the Akaike Information Criterion (corrected for finite sample size; AICc). For ResistanceGA, the AICc value is the fitness function output used for iteratively improving the relation between resistance surfaces and the response through the genetic algorithm. In addition, the MLPE allows accounting for the non-independence of values within pairwise distance matrices (Clarke et al. 2002; van Strien et al. 2012) and was fitted using maximum likelihood with the LME4 package (Bates et al. 2015).
The algorithm implemented in R package ResistanceGA roughly follows these steps in an iterative fashion: (i) least cost path calculation, based on each hypothesized surface, is performed to determine the landscape-based distance between each subpopulation (i.e., cost-distance matrix); (ii) the MLPE model is used to relate the genetic distance matrix (i.e., FST, G’’ST, and DST as the response) to the cost-distance matrix (i.e., predictor); (iii) from the model in step (ii) AICc is calculated to assess the level of support of each landscape surface; (iv) mathematical transformations are iteratively applied to the original surface, steps (i) to (iii) are recalculated, and each surface is compared and optimized through the AIC to achieve the best-performing resistance surface (i.e., by maximizing the explanatory power of the MLPE model on step (ii)).
Pairwise genetic distances FST, G’’ST, and DST are the response variables, while optimized cost-based resistance distances between populations are independent/predictor variables. The genetic algorithm then determines the best parameters for transforming the original resistance surface, optimising AICc model performance. Stopping criteria were set to a maximum of 250 rounds or 20 rounds without performance improvement.
To assess model performance, we compared AICc values between models generated for each optimized resistance surface to two baseline null models: (i) the isolation-by-distance (IBD) effect (which relies on the Euclidean distance between populations) and (ii) a neutral-landscape approach by simulating a gradient-like landscape (GL) using R package NLMR (Sciaini et al. 2018).
To determine which factors better explain gene flow, we ranked all models based on delta AICc values (ΔAICc). Models with ΔAICc ≤ 4 were considered with greater evidence/likelihood, thus entering the confidence set containing those models with substantial or good support for inference. We decided to use a less stringent threshold to identify potentially significant landscape-related factors explaining connectivity that would be missed if ΔAIC < 2 was used. We also calculated the Akaike weights (wi, between 0 and 1) to characterize the evidence strength of each model quantitatively.
Results
We amplified samples with a success rate of > 99% for both species. Allele dropouts or stuttering were detected in loci Aobstμ05 and Aobstμ16 in several Alytes populations. In Pelodytes populations, potential null alleles were only detected in two populations for one locus in each of them, P2 (Ppu10) and P7 (Ppu11).
We did not detect significant deviations from HWE and LD in Alytes, except in loci Aobstμ05 (A4, A5, A13, A16) and Aobstμ16 (A6, A7, A11, A14, A15), and between Aobstμ09 and Aobstμ17 (A2, A6), respectively. In Pelodytes, deviations from HWE were detected in Ppu5 (P11), Ppu8 (P10), Ppu9 (P10), Ppu10 (P13, P16), Ppu14 (P2, P13) and Ppu15 (P9, P16), whereas significant LD was found between loci Ppu8 and Ppu9 in several populations (P3, P4, P6, P10, P12, P13, P14, P16).
Descriptive statistics of genetic diversity for P. punctatus and A. obstetricans are presented in Table 1. Estimates of genetic diversity were slightly higher in P. punctatus. The mean number of alleles per population ranged from 2.6 (P16) to 9.7 (P9) in P. punctatus and from 2.9 (A16) to 7.8 (A9) in A. obstetricans. The observed heterozygosity ranged from 0.46 (P14) to 0.83 (P3) in P. punctatus, and from 0.44 (A16) to 0.75 (A9) in A. obstetricans. Average population inbreeding coefficients (FIS) were higher in A. obstetricans (0.03) than in P. punctatus (− 0.12).
The results of BAPS analyses supported optimal clustering levels at K = 16 and K = 13 for P. punctatus and A. obstetricans, respectively (Fig. 2). The number of clusters was consistent across replicate runs.
Optimal number of genetic clusters for Pelodytes punctatus (A) and Alytes obstetricans (B) according to BAPS analyses. Polygons represent Voronoi tessellations produced by the spatial clustering module in BAPS; each cell corresponds to the physical neighborhood of an observed data point, and is colored according to genetic cluster membership
Pairwise estimates of FST, G’’ST, and DST between species populations are presented in Supplementary material Tables S4-S9. Similar but slightly lower average values, indicating higher gene flow and greater population connectivity, were found in A. obstetricans (FST = 0.091; G’’ST = 0.264; DST = 0.209) than in P. punctatus (FST = 0.094; G’’ST = 0.326; DST = 0.260). Estimates of migration rates with BAYESASS were low across sites, except among geographically close localities (Supplementary material Tables S10 and S11), with slightly higher average migration rates between populations of A. obstetricans (mean = 0.0173) than in populations of P. punctatus (mean = 0.0133).
Resistance surfaces and gene flow
Overall, model ranking based on AICc and optimized resistance surfaces (Tables 3 and 4, and Supplementary material Table S12) showed a high degree of similarity for both species regarding the variables selected across different genetic distances.
Models for both species attained reasonably good performance values, as evidenced by the low rank of both null-models tested, the IBD and the simulated neutral GL. For A. obstetricans, a ΔAICc > > 4 denotes no support for null-models, ranging from ΔAICc = 19.96 for GL/FST to ΔAICc = 29.33 for IBD/G’’ST, (Table 3 and Supplementary material Table S12). For P. punctatus, despite slightly higher model uncertainty, null-models also attained no support, with ΔAICc = 11.25 for IBD/G’’ST to ΔAICc = 15.95 for GL/FST (Table 4 and Supplementary material Table S12). Marginal R2 values for selected models (ΔAICc ≤ 4) ranged from 0.22 to 0.38 for A. obstetricans and from 0.06 to 0.47 for P. punctatus.
Overall, distance to water lines and water availability are the most important features for both species, especially in A. obstetricans, as evidenced by the selection of variables related to the distance to rivers/wetlands or topographic water accumulation (Tables 3 and 4). Secondly, in terms of explanatory importance, is land cover/use, specifically (and in decreasing order of importance) the distance and the amount of artificial areas (housing and other types including roads, although with less spatial detail in comparison to road maps—See Fig. 3b, f), meadows and pastureland, and, with substantially less model support, agricultural areas, forest and heathland/shrubland (Supplementary material Table S13).
Transformations applied to selected resistance surfaces (i.e. with greater model support) based on ResistanceGA results. Top labels indicate the species, resistance surface and the genetic distance metric with best results in the analysis. The x-axis (“Original data values”) represents the data in the original scale/units while the y-axis (“Transformed data values”) shows the final resistance values after applying the optimized transformation. Top and lateral bars depict the histogram of each variable distribution
In general, distance-based resistance surfaces obtained greater explanatory importance than those depicting the percentage cover of different land use types (or their presence/absence) and continuous remote sensing indices for vegetation and water. This result suggests that the proximity to particular landscape features is generally more relevant than the composition of landscape mosaics in explaining gene flow among populations.
Contrary to our expectations (see e.g. Gutiérrez-Rodríguez et al. 2017), finer-scale features captured by satellite Earth Observations related to vegetation and water availability (in terms of amount or spatial heterogeneity) did not comparatively attain relevant importance in the models of both species. In line with this, layers including linear elements like roads or motorways did not attain much explanatory power either (Tables 3 and 4, and Supplementary material Table S12). Nonetheless, the distance to artificial/urban areas was highly relevant for Alytes obstetricans, supporting a major role of the whole network of anthropogenic structures driving landscape resistance.
Despite some similarities, there are strong contrasts when comparing the relative importance of different explanatory variables between species and how these translate into landscape resistance to movement (Figs. 3 and 4). In A. obstetricans up to three variables entered the confidence set, including Distance to water/wetlands, Distance to all artificial surfaces, and Distance to urban habitational areas (Table 3). In P. punctatus a different set of variables was selected, including Distance to meadows/pasturelands, Topographic Wetness Index, and River density -all rivers at 2500 m (Table 4).
Pairwise maps showing the original (blue) and transformed resistance surfaces (orange) through genetic algorithms optimization (based on G’’ST). Labels indicate the species and the resistance surface for: a A. obstetricans/Distance to water/wetlands, b A. obstetricans/Distance to all artificial surfaces, c P. punctatus/River density—all rivers at 2500 m, d P. punctatus/Topographic Wetness Index, e P. punctatus/Distance to meadows and pasturelands, f P. punctatus/Distance to urban/artificial areas
For A. obstetricans, the distance to water surfaces or lines is highly important for explaining gene flow, with resistance increasing non-linearly with increased distance to these areas (Figs. 3a and 4a). For P. punctatus, water is also important; however, its role manifests more in the density of the river network throughout the landscape, with resistance peaking at densities both very-low (absence of watercourses) and very-high (large rivers) (Figs. 3c and 4c). A minimum of resistance occurs for intermediate density of rivers and moderate wetness conditions as depicted by the topographic wetness index (Figs. 3d and 4d).
Overall, for A. obstetricans, distance and spatial distribution of artificial land-use types significantly influenced gene flow and connectivity patterns. In fact, the species seems to tolerate well urban/artificial elements in surrounding areas, with somewhat low resistance for smaller distances (i.e. within or close to urban/artificial patches) and increasing with greater distances to these landscape features (Figs. 3b and 4b). Still, given the widespread presence of artificial structures throughout the landscape and the interspersion of anthropogenic land uses, large distances to artificial areas/lines are not often encountered in the study area (see lateral histogram for the x-axis in Fig. 3b). In contrast, for P. punctatus, artificial/urban landscape elements have less importance, but their effect is also opposed to that in A. obstetricans. For P. punctatus, higher resistance is found within or in the vicinity of urban/artificial areas, decreasing progressively towards a minimum of resistance around 1000 m from artificial areas (Figs. 3f and 4f). Also, in contrast with Alytes, the amount and spatial distribution of meadows and pastureland areas also seem to shape gene flow in P. punctatus, with resistance peaking at small distances (within or in the vicinity ~ 0-500 m) to these land use patches (Figs. 3e and 4e) and with a minimum of resistance for a distance range of 700-900 m. Moreover, for G’’ST (Table 4), other LUC types (forest and heathland/shrubland) as well as elevation complexity (slope and TRI) also attained some explanatory power (although low) for P. punctatus, showing that a more extensive set of factors shape gene flow patterns in this species.
Discussion
We investigated the role of landscape features in shaping patterns of functional connectivity of two Mediterranean amphibian species in central Spain's semi-arid landscapes, where artificial irrigation structures provide critical breeding sites for some species. Understanding the role of different features in promoting or restricting gene flow is crucial to designing efficient management programs promoting the survival of amphibian communities in rural areas. Overall, our results provide evidence for higher population connectivity in common midwife toad (Alytes) than in common parsley frog (Pelodytes), with different landscape features shaping gene flow patterns for the two species. These differences should be considered when designing conservation actions targeted at each species separately, as suggested by studies on their breeding preferences (Caballero-Díaz et al. 2020). However, it should be kept in mind that our study is correlative, and while it provides an empirical approach to infer landscape factors likely impacting connectivity in our target species, the actual factors and processes involved are unknown. Experimental approaches investigating species responses to landscape elements would provide valuable information but at present remain challenging in wild amphibian populations.
At the regional scale, our comparative study showed marked genetic structure in both species (Fig. 2). Clustering analyses detected a higher number of genetic clusters in Pelodytes than in Alytes, which showed lower DST/FST/G’’ST values (Supplementary material Tables S4 to S9) and overall higher migration rates (mean = 0.0173 vs 0.0133 in Pelodytes). Unfortunately, there is little information about the dispersal capacity of the two species (Ryser et al. 2003; Trochet et al. 2014) as for most amphibians (Pittman et al. 2014). Based on indirect (molecular) evidence, Alytes and Pelodytes seem to be poor dispersers, which could make them vulnerable to habitat fragmentation processes. This hypothesis needs to be further tested, for instance, with information on the frequency and distances covered by adult individuals of both species in capture-mark-recapture studies.
Landscape genetic analyses provide evidence for similarities and differences in how the two species interact with the landscape, based on resistance values of different habitat feature layers (Tables 3 and 4). Our criterion to consider models with ΔAICc ≤ 4 is permissive, and inferences based on models with ΔAICc values ≥ 2 ≤ 4, while providing better explanatory power than null models, should be taken with caution. In general, distance to some landscape features showed greater explanatory power than features based on the composition of landscape mosaics. The influence of landscape features on population connectivity at the regional scale revealed a strong effect of water availability (Tables 3 and 4). Both the distance to rivers/wetlands and the topographic water accumulation index are key variables explaining functional connectivity in Alytes and Pelodytes. This dependence on the presence of water is expected in Mediterranean semi-arid landscapes, where animal and plant communities are constrained by water availability (Noy-Meir 1973; Dodd and Lauenroth 1997). This effect is clearly the case in our study area, with an annual average precipitation of 415 mm, and almost no rainfall during the summer (between 250 and 600 mm; Romão and Escudero 2005; García et al. 2011). Climatic models have predicted a significant decrease in water availability in the Mediterranean basin (Houghton et al. 2001; Polade et al. 2017), which may have dramatic consequences for water-dependent species (Bates et al. 2008).
Beyond their shared dependence on water availability, our models provide evidence that landscape variables played a differential role on patterns of population connectivity in both species. In P. punctatus, the absence of watercourses and the presence of large rivers restricted gene flow among populations (Figs. 3c and 4c), with genetic connectivity increasing at intermediate river densities (Figs. 3d and 4d). In contrast, in A. obstetricans, distance to water shaped gene flow differently, with resistance increasing non-linearly with increased distance to these areas (Figs. 3a and 4a). These differences are probably associated with differences in the reproductive biology of the two species. The common parsley frog is very flexible in using aquatic habitats, usually occupying shallow temporary ponds in meadows and pastures for reproduction (Guyétant et al. 1999; Boix et al. 2001; Grillas et al. 2004; Salvidio et al. 2004; Tatin 2010; Escoriza 2017). In contrast, common midwife toads tend to choose long-hydroperiod or permanent water bodies because of their longer larval stage (Bosch et al. 2009). In the study area, Caballero-Díaz et al. (2020) found marked differences in the selection of breeding sites between the two species, with Alytes preferring artificial water bodies (fountains, water tanks) and Pelodytes breeding mostly in temporary ponds.
A second important difference is the role of variables related to land cover/use on patterns of gene flow in the two species. The distance and spatial distribution of artificial land-use types positively influenced connectivity in A. obstetricans, which seems to tolerate the presence of urban/artificial elements well. This effect is probably related to their ability to exploit artificial breeding sites successfully, many of which are located in peri-urban settings, including orchards and recreational areas (Caballero-Díaz et al. 2020). In contrast, in P. punctatus, which rarely breeds in those artificial water structures in urban areas, the amount and spatial distribution of meadows and pastureland areas were important positive predictors of gene flow.
This study highlights the importance of assessing species–habitat relationships shaping gene flow and population connectivity when developing and implementing conservation and management actions to benefit fragmented amphibian populations in the Mediterranean region. As previously shown for other co-distributed Mediterranean amphibians (Gutiérrez-Rodríguez et al. 2017), our results show that amphibian species respond differently, even contrastingly to landscape features and thus require alternative, complementary strategies to improve population connectivity and ensure their long-term viability. For instance, the construction and maintenance of artificial ponds can be an efficient resource to create and sustain viable, well-interconnected populations of common midwife toads (Alytes), whereas measures directed to improve the conservation status of Pelodytes populations should focus on the conservation of natural habitats in low-resistance areas, including meadows and floodable pasturelands.
References
Auffret AG, Plue J, Cousins SA (2015) The spatial and temporal components of functional connectivity in fragmented landscapes. Ambio 44(Suppl 1):S51-59
Bates B, Kundzewicz Z, Wu S (2008) Climate change and water. Intergovernmental Panel on Climate Change Secretariat, Geneva
Bates D, Maechler M, Bolker B, Walker S (2015) Fitting linear mixed-effects models using lme4. J Stat Softw 67(1):1–48
Boix D, Sala J, Moreno-Amich R (2001) The faunal composition of Espolla pond (NE Iberian Peninsula): the neglected biodiversity of temporary waters. Wetlands 21:577–592
Bosch J, Beebee T, Schmidt B, Tejedo M, Martínez-Solano I, Salvador A, García-París M, Recuero Gil E, Arntzen JW, Díaz Paniagua C, Márquez R (2009) Alytes obstetricans. IUCN Red List Threatened Species 2009: e.T55268A11283700
Bouahim S, Rhazi L, Amami B, Sahib N, Rhazi M, Waterkeyn A, Zouahri A, Mesleard F, Muller SD, Grillas P (2010) Impact of grazing on the species richness of plant communities in Mediterranean temporary pools (western Morocco). C R Biol 333(9):670–679
Caballero-Díaz C, Sánchez-Montes G, Butler HM, Vredenburg VT, Martínez-Solano I (2020) The role of artificial breeding sites in amphibian conservation: a case study in rural areas in central Spain. Herpetol Conserv Biol 15(1):87–104
Cayuela H, Besnard A, Bechet A, Devictor V, Olivier A (2012) Reproductive dynamics of three amphibian species in Mediterranean wetlands: the role of local precipitation and hydrological regimes. Freshw Biol 57(12):2629–2640
Cheng L, Connor TR, Sirén J, Aanensen DM, Corander J (2013) Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol Biol Evol 30(5):1224–1228
Clarke RT, Rothery P, Raybould AF (2002) Confidence limits for regression relationships between distance matrices: estimating gene flow with distance. Agric Biol Environ Stat 7(3):361–372
Corander J, Sirén J, Arjas E (2008) Bayesian spatial modeling of genetic population structure. Comput Stat 23(1):111–129
Crawford NG (2010) SMOGD: software for the measurement of genetic diversity. Mol Ecol Resour 10(3):556–557
Cuttelod A, García N, Malak DA, Temple HJ, Katariya V (2009) The Mediterranean: a biodiversity hotspot under threat. In: Vié JC, Hilton-Taylor C (eds) Wildlife in a Changing World–an analysis of the 2008 IUCN Red List of Threatened Species. IUCN, Gland, p 89
Denoël M, Beja P, Andreone F, Bosch J, Miaud C, Tejedo M, Lizana M, Martínez-Solano I, Salvador A, García-París M, Recuero Gil E, Márquez R, Cheylan M, Díaz-Paniagua C, Pérez-Mellado V (2009) Pelodytes punctatus. IUCN Red List Threatened Species 2009: e.T58056A11710052
Dodd MB, Lauenroth WK (1997) The influence of soil texture on the soil water dynamics and vegetation structure of a shortgrass steppe ecosystem. Plant Ecol 133(1):13–28
Escoriza D (2017) Sapillo moteado septentrional—Pelodytes punctatus. In: Sanz JJ, Martínez-Solano I (eds) Enciclopedia Virtual de los Vertebrados Españoles. Museo Nacional de Ciencias Naturales, Madrid
García R, González JA, Rubio V, Arteaga C, Galán A (2011) Soil contamination in dumps on the karstic areas from the plateaus (Southeast of Madrid, Spain). Water Air Soil Pollut 222(1):27–37
Gómez-Rodríguez C, Bustamante J, Díaz-Paniagua C (2010a) Evidence of hydroperiod shortening in a preserved system of temporary ponds. Remote Sens 2(6):1439–1462
Gómez-Rodríguez C, Díaz-Paniagua C, Serrano L, Florencio M, Portheault A (2009) Mediterranean temporary ponds as amphibian breeding habitats: the importance of preserving pond networks. Aquat Ecol 43(4):1179
Gómez-Rodríguez C, Díaz-Paniagua C, Bustamante J, Portheault A, Florencio M (2010b) Inter-annual variability in amphibian assemblages: implications for diversity assessment and conservation. Aquat Conserv: Mar Freshw Ecosyst 20(6):668–677
Gómez-Rodríguez C, Díaz-Paniagua C, Bustamante J, Serrano L, Portheault A (2010c) Relative importance of dynamic and static environmental variables as predictors of amphibian diversity patterns. Acta Oecol 36(6):650–658
Gonçalves J, Honrado JP, Vicente JR, Civantos E (2016) A model-based framework for assessing the vulnerability of low dispersal vertebrates to landscape fragmentation under environmental change. Ecol Complex 28:174–186
Green AJ, El Hamzaoui M, El Agbani MA, Franchimont J (2002) The conservation status of Moroccan wetlands with particular reference to waterbirds and to changes since 1978. Biol Conserv 104(1):71–82
Grillas P, Gauthier P, Yavercovsky N, Perennou C (2004) Les mares temporaires méditerranéennes. Volume 1—Enjeux de conservation, fonctionnementet gestion. Station Biologique Tour de Valat, Arles
Gutiérrez-Rodríguez J, Gonçalves J, Civantos E, Martínez-Solano I (2017) Comparative landscape genetics of pond-breeding amphibians in Mediterranean temporal wetlands: the positive role of structural heterogeneity in promoting gene flow. Mol Ecol 26(20):5407–5420
Guyétant R, Temmermans W, Avrillier JN (1999) Phénologie de la reproduction chez Pelodytes punctatus Daudin, 1802 (Amphibia, Anura). Amphib-Reptilia 20:149–160
Houghton JT, Ding YDJG, Griggs DJ, Noguer M, van der Linden PJ, Dai X, Maskell K, Johnson CA (2001) Climate change 2001: the scientific basis. The Press Syndicate of the University of Cambridge, Cambridge
Izco J (1984) Madrid Verde. Ministerio de Agricultura, Pesca y Alimentación, Madrid
Jakob C, Poizat G, Veith M, Seitz A, Crivelli AJ (2003) Breeding phenology and larval distribution of amphibians in a Mediterranean pond network with unpredictable hydrology. Hydrobiologia 499(1–3):51–61
Jost L (2008) GST and its relatives do not measure differentiation. Mol Ecol 17(18):4015–4026
Jost L, Archer F, Flanagan S, Gaggiotti O, Hoban S, Latch E (2018) Differentiation measures for conservation genetics. Evol Appl 11(7):1139–1148
Jourdan-Pineau H, Nicot A, Dupuy V, David P, Crochet PA (2009) Development of eight microsatellite markers in the parsley frog (Pelodytes punctatus). Mol Ecol Resour 9(1):261–263
Keddy PA (2010) Wetland ecology: principles and conservation. Cambridge University Press, New York City
Lourenço A, Gonçalves J, Carvalho F, Wang IJ, Velo-Antón G (2019) Comparative landscape genetics reveals the evolution of viviparity reduces genetic connectivity in fire salamanders. Mol Ecol 28(20):4573–4591
Maia-Carvalho B, Gonçalves H, Martínez-Solano I, Gutiérrez-Rodríguez J, Lopes S, Ferrand N, Sequeira F (2014) Intraspecific genetic variation in the common midwife toad (Alytes obstetricans): subspecies assignment using mitochondrial and microsatellite markers. J Zool Syst Evol Res 52(2):170–175
Martínez-Solano I (2006) Atlas de distribución y estado de conservación de los anfibios de la Comunidad de Madrid. Graellsia 62(Extra):253–291
Médail F, Quézel P (1999) Biodiversity hotspots in the Mediterranean Basin: setting global conservation priorities. Biol Conserv 13(6):1510–1513
Meirmans PG, Hedrick PW (2011) Assessing population structure: FST and related measures. Mol Ecol Resour 11(1):5–18
Meirmans PG, van Tienderen PH (2004) GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes 4(4):792–794
Newman RA (1992) Adaptive plasticity in amphibian metamorphosis: what type of phenotypic variation is adaptive, and what are the costs of such plasticity. Bioscience 42(9):671–678
Noy-Meir I (1973) Desert ecosystems: environment and producers. Annu Rev Ecol Evol Syst 4(1):25–51
Parmesan C (2006) Ecological and evolutionary responses to recent climate change. Annu Rev Ecol Evol Syst 37:637–669
Peakall R, Smouse PE (2012) GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28(19):2537–2539
Pearce F, Crivelli AJ (1994) Characteristics of Mediterranean wetlands. In: Crivelli AJ, Jalbert J (eds) Conservation of Mediterranean Wetlands 1. MedWet publications, Station Biologique de la Tour du Valat, Arles
Pérez-Espona S, Pérez-Barbería FJ, McLeod JE, Jiggins CD, Gordon IJ, Pemberton JM (2008) Landscape features affect gene flow of Scottish Highland red deer (Cervus elaphus). Mol Ecol 17(4):981–996
Peterman WE (2018) ResistanceGA: An R package for the optimization of resistance surfaces using genetic algorithms. Methods Ecol Evol 9(6):1638–1647
Peterman WE, Connette GM, Semlitsch RD, Eggert LS (2014) Ecological resistance surfaces predict fine-scale genetic differentiation in a terrestrial woodland salamander. Mol Ecol 23(10):2402–2413
Pittman SE, Osbourn MS, Semlitsch RD (2014) Movement ecology of amphibians: a missing component for understanding population declines. Biol Conserv 169:44–53
Polade SD, Gershunov A, Cayan DR, Dettinger MD, Pierce DW (2017) Precipitation in a warming world: assessing projected hydro-climate changes in California and other Mediterranean climate regions. Sci Rep 7(1):1–10
Quinn PF, Beven KJ, Lamb R (1995) The in(a/tan/β) index: How to calculate it and how to use it within the topmodel framework. Hydrol Process 9(2):161–182
R Core Team (2020) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA (2018) Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst Biol 67(5):901–904
Raymond M, Rousset F (1995) GENEPOP (Version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Rhazi L, Grillas P, Saber ER, Rhazi M, Brendonck L, Waterkeyn A (2012) Vegetation of Mediterranean temporary pools: a fading jewel? Hydrobiologia 689(1):23–36
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43(1):223–225
Romão RL, Escudero A (2005) Gypsum physical soil crusts and the existence of gypsophytes in semi-arid central Spain. Plant Ecol 181(1):127–137
Rousset F (2008) genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour 8(1):103–106
Ryser J, Lüscher B, Neuenschwander U, Zumbach S (2003) Geburtshelferkröten im Emmental. Schweiz Z Feldherpetol 10:27–35
Sahuquillo M, Miracle MR, Morata SM, Vicente E (2012) Nutrient dynamics in water and sediment of Mediterranean ponds across a wide hydroperiod gradient. Limnologica 42(4):282–290
Salvidio S, Lamagni L, Bombi P, Bologna MA (2004) Distribution, ecology and conservation of the parsley frog (Pelodytes punctatus) in Italy (Amphibia, Pelodytidae). Boll Zool 71:73–81
Sciaini M, Fritsch M, Scherer C, Simpkins CE (2018) NLMR and landscapetools: An integrated environment for simulating and modifying neutral landscape models in R. Methods Ecol Evol 9(11):2240–2248
Storfer A, Murphy MA, Evans JS, Goldberg CS, Robinson S, Spear SF, Dezzani R, Delmelle E, Vierling L, Waits LP (2007) Putting the ‘landscape’in landscape genetics. Heredity 98(3):128–142
Stuart SN, Chanson JS, Cox NA, Young BE, Rodrigues AS, Fischman DL, Waller RW (2004) Status and trends of amphibian declines and extinctions worldwide. Science 306(5702):1783–1786
Stuart SN, Hoffmann M, Chanson JS, Cox NA, Berridge RJ, Ramani P, Young BE (2008) Threatened amphibians of the world. Lynx Edicions, Barcelona
Tatin D (2010) Les mares et amphibiens de la vallée du Calavon et du pays d’Apt: étude et premières mesures de gestion conservatoire. Courrier Sci Parc Nat Rég Luberon 9:88–100
Trochet A, Moulherat S, Calvez O, Stevens V, Clobert J, Schmeller D (2014) A database of life-history traits of European amphibians. Biodivers Data J 2:e4123
van de Vliet MS, Diekmann OE, Serrao ET, Beja P (2009) Highly polymorphic microsatellite loci for the Parsley frog (Pelodytes punctatus): characterization and testing for cross-species amplification. Conserv Genet 10(3):665–668
van Etten J (2017) R package gdistance: distances and routes on geographical grids. J Stat Softw 76(1):1–21
van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4(3):535–538
van Strien MJ, Keller D, Holderegger R (2012) A new analytical approach to landscape genetic modelling: least-cost transect analysis and linear mixed models. Mol Ecol 21(16):4010–4023
Vanschoenwinkel B, Hulsmans ANN, De Roeck E, De Vries C, Seaman M, Brendonck L (2009) Community structure in temporary freshwater pools: disentangling the effects of habitat size and hydroregime. Freshw Biol 54(7):1487–1500
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38(6):1358–1370
Williams DD (2006) The biology of temporary waters. Oxford University Press, Oxford
Wilson GA, Rannala B (2003) Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163(3):1177–1191
Acknowledgements
We thank E. Ayllón, E. González, P. Hernández, S. Jiménez, L. Martín, B. Paños, E. Recuero, M. Sánchez and I. Urbán for help during field work or for providing samples.
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature. JGR was supported by “Doctores Junta de Andalucía” postdoctoral fellowship (DOC_00668, FEDER EU/Consejería de Economía, Conocimiento, Empresas y Universidad, Junta de Andalucía). JG was funded by the Individual Scientific Employment Stimulus Program (2017) by the Portuguese Foundation for Science and Technology (FCT CEECIND/02331/2017/CP1423/CT0012). EC was supported by the FCT through a Postdoctoral Grant (SFRH/BPD/109182/2015). This study was funded by projects CGL2008-04271-C02-01/BOS, CGL2011-28300 (Ministerio de Ciencia e Innovación, Ministerio de Economía y Competitividad, Spain, and FEDER), PPII10-0097-4200 (Junta de Comunidades de Castilla la Mancha and FEDER), and CGL2017-83131-P (FEDER/Ministerio de Ciencia, Innovación y Universidades–Agencia Estatal de Investigación, Spain).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gutiérrez-Rodríguez, J., Gonçalves, J., Civantos, E. et al. The role of habitat features in patterns of population connectivity of two Mediterranean amphibians in arid landscapes of central Iberia. Landsc Ecol 38, 99–116 (2023). https://doi.org/10.1007/s10980-022-01548-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10980-022-01548-z