Abstract
Context
Disentangling the effect of environment and biological interaction on community composition with observational data, within the environmental filtering framework, is challenging because the two processes produce non independent results.
Objectives
Adopting community N-mixture models with symmetric interactions, we aimed at estimating differential effects of landscape structure and biotic interactions on the local abundance of a Mediterranean reptile community including four lizards (Lacerta bilineata; Podarcis siculus; P. muralis; Chalcides chalcides) and two snakes (Hierophis viridiflavus; Natrix Helvetica).
Methods
We sampled reptiles for three consecutive years (2019–2021; 4 surveys/year) on 52 linear transects on a Mediterranean coastal landscape. We analyzed count data by means of a multi-species N-mixture model with symmetric interactions. Interactions within pair of species were estimated from the residual correlation of their realized abundances, after accounting for four landscape features: landscape heterogeneity calculated from land cover data, edge density of woody vegetation patches, tree cover density, net primary productivity.
Results
Most species displayed very low detection probability (p ~ 0.10). All species responded with different intensity and sensitivity to landscape predictors. Two biological interactions resulted significant: L. bilineata and P. siculus showed a positive interaction, while P. muralis and C. chalcides displayed a negative interaction.
Conclusions
Using community N-mixture models we demonstrated that, also with observational data obtained from a realized community, partitioning the filtering process of the landscape from the one of biotic interactions is possible.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Community composition and local abundance of species, at the landscape scale, are shaped by both environmental features and biotic interactions occurring among community members. The effect of abiotic environment and biotic interactions on community assembly has been largely studied within the framework of environmental filtering (Kraft et al. 2015). This conceptual framework suggests that environmental constraints and biotic interactions contribute to select ecological traits, filtering out species with less adapted traits for a given condition, thus determining local extinctions (Kraft et al. 2015; Germain et al. 2018). These filtering effects are supposed to act on a hierarchical level, and at different spatial and temporal scales: from higher processes of regional extinction and dispersal, occurring at evolutionary timescales (Ricklefs 2004), to lower processes of environmental (e.g. landscape features) and biotic filtering (e.g. inter-specific competition or predation), that shapes species composition and abundances of local populations within a realized community, living in a given location or landscape patch (Keddy 1992; Fig. 1). This classical, well-established, heuristic approach extended toward 2000’s (e.g. Swenson et al. 2012; Whitfeld et al. 2012). However, in recent years some ecologists gave rise to a debate on the viability of the environmental filtering concept. In particular, the usefulness of observational studies in disentangling the effect of environment and within-community biotic interactions has been questioned (Kraft et al. 2015; Cadotte and Tucker 2017). This is because the ecological effects of both competition between community members and the influence of environment on species occurrence and abundance are not independent, and observational studies preclude investigation into their interplay (Kraft et al. 2015). This debate expanded the field of application of environmental filtering concept (e.g. Gearty et al. 2021), boosting research in novel approaches capable of disentangling these ecological processes (e.g. Germain et al. 2018).
In this context, the application of Hierarchical Models—i.e. Occupancy and N-mixture models (see for instance Kéry and Royle 2015, 2020)—seems promising to better disentangle the effect of environment and biotic interactions within communities exposed to different ecological gradients or anthropogenic disturbances (Brodie et al. 2017; Montano-Centellas et al. 2021; Basile et al. 2021; Rosa et al. 2022a), because these models can be fit to a variety of ecological process (Kéry and Royle 2015, 2020) and account for imperfect detection (i.e. the difficulty of observing rare and secretive species). In fact, community Occupancy and N-mixture models have been recently developed to account for biotic interactions occurring between species and allow to disentangle the contribution of the environment from the one of biotic interactions on occurrence and local abundance of each species (see Kéry and Royle 2020 for a detailed discussion). These models can be divided in two groups: (i) models accounting for direct biotic interactions in model structure, i.e. the realized abundance or occurrence of a species has a direct effect on the realized state of another species (Waddle et al. 2010; Clare et al., 2016; Brodie et al. 2018), and (ii) models that account for symmetric interactions between species, meaning that the effect of biotic interaction is not included in model structure, but is inferred by means of residual correlation between the realized state of two species (i.e. abundance or occurrence; Pollock et al. 2014; Tobler et al. 2019). Both models, with direct and symmetric interactions, are effective to infer species interaction for both complex and reduced species pools: however, models with direct interactions are usually employed when few species are present and a clear effect of one species on another is assumed a priori (e.g. Brodie et al. 2018; Rosa et al. 2022a). Conversely, models with symmetric interaction are usually adopted when many species are analyzed or when direction of biotic interaction cannot be a priori hypothesized (but see Basile et al. 2021). Therefore, these models represent a promising and innovative framework to understand the determinants of community composition and structure, and to quantify the hierarchical processes of environmental and biotic filtering on the observed local biological communities (Kéry and Royle 2020; Ovaskainen and Abrego 2020). Specifically, significant species interactions observed after accounting for environmental filtering, can be attributable to different ecological processes, depending on the trophic level of the community members: (i) inter-specific competition for resources should result in negative interactions between species of the same trophic level, (ii) interactions occurring between species at different trophic levels, mediated by predator–prey dynamics, could both result in positive abundance interactions, when predators’ pressure is low or prey density is high, or negative abundance interactions when predators’ pressure is more pronounced (Chase et al. 2002).
The Mediterranean global biodiversity hotspot (Myers et al. 2000) is recognized as an example of human-modified dynamic landscape, constantly shaped for millennia by long-lasting and constantly evolving land use practices (Blondel and Aronson 1999). Indeed, human activities in the Mediterranean basin have determined a dynamic mosaic of different habitats and vegetation, such as scrublands, forests, and agricultural patches (Blondel 2006). Within Europe this heterogeneous landscape, where patches of different natural vegetation and agricultural land-use coexist at a small scale, is capable of maintaining high levels of both taxonomic and functional biodiversity (e.g. Burel and Baudry 1995; Potts et al. 2006; Sirami et al. 2010). For this reason, ecologists historically spent great efforts to understand the ecological processes determining biodiversity patterns within the Mediterranean region. In particular, they were intrigued by biological community assembly rules at landscape scale: considering interactive effects of historical land-use practices (e.g. Ruiz 1990; Farina 1997), patchy human perturbances such as fires (e.g. Brotons et al. 2005; Santos and Cheylan, 2013), and more recently by considering the effects of regionally diffuse dynamics, such as land-abandonment (Plieninger et al. 2014), climate change and diffusion of pathogens (e.g. Casazza et al. 2021).
Among the many biological communities exposed to anthropogenic landscape changes in the Mediterranean region, Reptiles represent one of the most endangered class of vertebrates (Cox et al. 2022). The effects of anthropogenic pressures, such as forest fires and intensive land use practices, on reptile populations and communities is therefore well studied (e.g. Ribeiro et al. 2009; Rotem et al. 2013; Guiller et al. 2022). However, a comprehensive evaluation of the effects of environmental features and biotic interactions in Mediterranean human-modified landscapes is poorly understood. In this study we aim at using observational data from a realized community at the landscape scale to evaluate if, once landscape metrics are accounted for, species interactions can still influence local abundance. Adopting community N-mixture models with symmetric interactions, we estimated differential effects of landscape and biotic interactions on the composition and local abundance of a Mediterranean reptile community, focusing on landscape metrics in an area where both natural, semi-natural and human-modified environments are present. Moreover, we also aim at inferring landscape requirements of different species within the community, to properly inform local conservation programs and landscape management plans.
Methods
Study rationale and conceptual framework
In the present study we aimed at investigating landscape-scale determinants of the local abundance of a Mediterranean reptile community, framing community assembly processes within the environmental filtering conceptual model (Keddy 1992). In our study we considered a reptile community in central Italy, which we repeatedly sampled in 52 sites, covering about 60 km2 of a Mediterranean coastal and subcoastal environment, consisting of a mosaic of natural and human-modified habitat patches. In particular, we adopted recently developed community N-mixture models, which take into account species interactions (Kéry and Royle 2020), to decouple the two hierarchies of the filtering process: environmental filtering by the landscape and biotic filtering occurring within community members (Fig. 1).
Study site
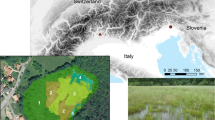
The study area (Fig. 2) is a costal and subcostal plane of about 20 × 3 km, from the sea level to 25 m a.s.l, from the Foglino forest and, southernward, up to the Fogliano Lake (Latium, Central Italy). This territory belongs to the widest area known as Pontine Plain which is the result of an extensive land reclamation of the Pontine swamps, conducted by Fascist regime between 1920 and 1936, that radically changed the landscape of the area. Different habitats and environments can be found: the Foglino forest (Fig. 2c) is a wide residual patch of the planitial and partially flooded forest dominated by deciduous oaks (last coppicing occurred in 90’s of the XX century; Lattanzi et al. 2004); other main habitat are patches of Pinus sp. plantation forests along a part of the retrodunal areas (Fig. (Fig. 2d); Mediterranean shrubs (Fig. 2e); agricultural environments, mainly for hay meadow (Fig. 2f) and open areas/pastures (Fig. 2g).
a Location of the study area in Italy; b detailed location of the study area on the Latium coastline; c–g habitats comprised within the study area, representative of the Mediterranean landscape: c Oak forests, d Pinus sp. plantation forests, e Mediterranean shrubs, f agricultural environments (hay meadow), g open areas/pastures
In the study area, 52 linear transects (about 200 × 4 m), approximately equally divided among these different environmental typologies (9–11 transect for each typology), were placed and surveyed repeatedly over three years.
Sampling
Sampling occurred during spring and early summer over three consecutive years. We visited each of the 52 transects 4 times both in 2020 and 2021. Surveys occurred between March 3rd and July 5th (both in 2019), while we conducted 3 visits on 47 transects in 2019. Two observers (R.N. and A.R.) slowly walked each 200-m transect line and conducted standardized visual encounter surveys, recording the number of reptile individuals encountered. Surveys took place during daytime (between 8.00 and 20.00) and lasted between 10 and 20 min, according to environmental complexity of the transect. For each survey observers recorded the time of the day (HOUR) and the day of the year (DAY; i.e. the continuous count of the number of days beginning each year from January 1st).
Landscape metrics
For each sampling site we calculated four landscape metrics, supposed to influence occurrence and abundance of reptiles (Atauri and De Lucio 2001; Schindler et al. 2013). Considering that environmental features in close proximity to the transect have an influence on the transect line itself, we applied a buffer of 20 m radius along each transect line, thus obtaining a rectangular polygon with rounded edges (240 × 40 m; length x width) covering an area of almost 1 ha, which we used to estimate landscape metrics for the analysis. Starting from the Corine Land Cover 2018 layer (CLC; 100 m spatial resolution; European Environment Agency 2019a) we calculated the number of CLC categories present on each polygon, which we assumed to represent landscape diversity or land use heterogeneity across each polygon (LUH). For each polygon, we extracted the value of the net primary productivity (NPP; expressed as kgC/m2/year) from the Annual Terra Moderate Resolution Imaging Spectroradiometer (MODIS) layer (500 m spatial resolution; Running and Zhao 2021) for a given year, averaging cell values when polygon fell within two cells. From the European Environment Agency (EEA) we obtained a high-resolution layer concerning vegetation tree cover density (TCD; 10 m spatial resolution; reference year 2018; European Environment Agency 2019b), ranging from 0 to 100%, from which we calculated the median value for each polygon. From the EEA we also obtained a high-resolution layer representing the presence/absence of small woody features (SWF; 5 m spatial resolution; reference year 2015; European Environment Agency 2019c), defined as hedgerows or bushes. Combining TCD and SWF we obtained an additional layer (5 m spatial resolution), representative of the presence/absence of arboreal or shrub vegetation. From this layer we calculated the edge density for each polygon (EDGE; expressed in m/ha), which gives, for each sampling site, a measure of ecotone complexity between woody vegetation and other environmental patches (Schindler et al. 2013). All spatial analyses and layer manipulations were conducted with free software QGIS v.3.22. Edge density and polygon median values were calculated with Landscape Ecology Statistics plugin (LecoS; Jung 2016) for QGIS.
Data analysis
We analysed our repeated count data of Mediterranean reptiles using a multi-species formulation of the static binomial N-mixture model of Royle (2004) accounting for symmetric interactions between community members (Pollock et al. 2014; Tobler et al. 2019). Symmetric interactions within pair of species were estimated from the residual correlation of their realized abundances, after accounting for the environmental filtering, i.e. the effect of landscape features (Kéry and Royle 2020). These interactions may be negative or positive according to the biological process underlying relationship between species (Kéry and Royle 2020). Binomial N-mixture models estimate latent abundance state N at site i (Ni), assuming Ni ~ Poisson(λ), where λ is the expected abundance over all sites, by using repeated counts C at site i during survey j (Cij) to estimate individual detection probability p, assuming Cij|Ni ~ Binomial(Ni,p). Both parameters can be modelled as a function of environmental covariates trough a log or logit link, respectively, which conveniently allowed us to include landscape metrics as covariates. In order to model species interactions, we included site- and species- specific random effects with a Multivariate Normal (MVN) distribution (Kéry and Royle 2020). Off-diagonal elements of the variance–covariance matrix of the MVN are scaled to obtain residual abundance correlations (ρ), which measure the direction and the intensity of the interaction of each species pair considered, after the effect of environmental covariates on abundance have been accounted for (Kéry and Royle 2020). Finally, we considered temporal dynamics in our dataset by fitting a year-stratified model (Kéry and Royle 2020), i.e. by stacking the data of subsequent years and treating them as different sites, so that site i in year 2019 and site i in year 2020 appear as different rows of the dataset. This approach allows to account for trends in the data by including a temporal covariate (YEAR). Prior to building our model, we standardized covariates and checked them for collinearity, considering a cut-off for inclusion of Pearson r < 0.7 (Dormann et al. 2013). We modelled the detection process of each species as follows:
where α0 is the intercept, α1- α2 are covariate effects and δ is a random effect, assuming normal distribution, to account for possible over dispersion in the detection process (Kéry and Schaub 2011). For the abundance of each species, we modelled the filtering effect of landscape metrics on the community as follows:
where for a given species, β0 is the intercept, β1- β5 are covariate effects to account for temporal dynamics and landscape environmental filtering, while η is the site- and species- specific random effects with Multivariate Normal (MVN) distribution included to measure biotic interactions (Kéry and Royle 2020). We estimated model parameters using a Bayesian approach with Markov chain Monte-Carlo methods, using un-informative priors. We ran three chains, each one with 550,000 iterations, discarding the first 120,000 as a burn-in and thinning by 100. We considered that chains reached convergence when the Gelman-Rubin statistic was < 1.1 (Gelman and Rubin 1992). We considered parameters relative to environmental covariates and species interactions to have a significant effect when the 90% Credible Interval (CRI) of the posterior distribution did not cross the zero. The N-mixture model used here is derived from the static binomial N-mixture model proposed by Royle (2004), and is prone to the same criticism on parameter identifiability, assumption violation and overdispersion (e.g. Barker et al. 2018; Link et al. 2018; but see Kéry et al. 2018). Because of this, properly assessing model fit is of primary importance (Duarte et al. 2018; Costa et al. 2020, 2021). Therefore, we utilized posterior predictive checks based on χ2 statistics as a measure of the discrepancy between observed and simulated data and calculated a Bayesian p-value accordingly (Kéry and Schaub 2011). In order to obtain reliable estimates of detection, abundance and biotic interactions, we applied a threshold to count data and retained for the analysis only those species with > 3 observations for at least one study year. Analyses were conducted calling program JAGS (V4.3.1; Plummer 2003) from the R environment with package “JagsUI” (V1.5.1; Kellner 2015). The JAGS code used for the analysis is available as Supplementary Information SI.1.
Results
During three years of sampling on our transects we encountered twelve reptile species. However, some species were only occasionally encountered (turtles: Emys orbicularis; lizards: Anguis veronensis, Hemidactylus turcicus; snakes: Elaphe quatuorlineata, Zamenis lineatus, Vipera aspis), and we retained for the analysis six species. We counted a total of 2270 Reptiles (Table 1) relative to: three Lacertid lizards (Lacerta bilineata, Podarcis siculus, Podarcis muralis), one Scincid lizard (Chalcides chalcides) and two Colubrid snakes (Hierophis viridiflavus, Natrix helvetica (species descriptions are available as Supplementary Information SI.2). After evaluating goodness-of-fit of our multi-species N-mixture model with symmetric interactions, by means of posterior predictive checks, we recorded a good fit for all species included in the analysis (Supplementary Information SI.3). Convergence, assessed by means of R-hat value, was successful for all parameters monitored (maximum R-hat = 1.040). Mean individual detection probability was very low (i.e., p ~ 0.10) for almost all species (Table 1), except for the most common Lacertid lizards P. siculus and P. muralis that displayed a slightly higher detection probability (i.e., p ~ 0.20). Moreover, for the majority of species detection probability resulted to be mainly influenced by phenology (DAY), and at a lesser extent by daily activity (HOUR; Supplementary Information SI.4). Mean site abundance greatly differed among species: the most abundant species within the community being P. siculus (λ = 8.27; 90% CRI = 6.63–10.49), while for other species we estimated less than 1 individual per transect on average (λ < 1; Table 1). For what concerns the effect of landscape features on the local abundance of species, all species responded with different sensitivity and intensity to the set of landscape predictors included in the model, and complete results are resumed in Fig. 3. For instance, the abundance of both L. bilineata and C. chalcides resulted to be negatively affected by NPP, but for the latter also a negative response of TCD was observed, while EDGE had a positive effect on abundance (Fig. 3). Similarly, the most commonly observed Lacertid lizards in the study area, P. siculus and P. muralis, had different sensitivity as well as opposite responses to some landscape scale predictors of abundance. That is, P. siculus abundance was negatively affected by TCD, while P. muralis was positively affected by the same predictor and at the same time negatively affected by NPP and LUH (Fig. 3). For neither of the snake species included in the analysis we recorded a significant effect of any landscape feature on local abundance (Fig. 3). Significant temporal dynamics in the year-stratified model (i.e., YEAR predictor on abundance) were observed for three species within the studied community: P. siculus and C. chalcides showed a positive temporal dynamic during the study period, while P. muralis showed a negative dynamic (Fig. 3). Finally, after accounting for the effect of environmental predictors on abundance and measuring correlation of residual abundances, we observed mixed biotic interactions across species pairs within the community (Fig. 4). Among all species pairs, only two accounted for a significant biological interaction: L. bilineata and P. siculus showed a positive interaction (ρ = 0.58; 90% CRI = 0.25–0.81), while P. muralis and C. chalcides displayed a strong negative interaction (ρ = − 0.76; 90% CRI = − 0.92 to − 0.39).
Plots representing posterior distributions of covariate effects for abundance predictors included in the model, for each species. Empty areas represent 100% of the posterior distribution, filled areas represent 90% CRI, bold lines within posterior distributions represent point estimates (mean) for a given parameter. When > 90% of the posterior distribution for a given predictor falls on one side of the 0 vertical line, the predictor of interest is considered to have a significant effect on the relative parameter
Plot representing biotic interaction coefficients for all species pairs: positive abundance interactions are depicted in blue, negative abundance interactions in red, color intensity represents interaction strength. Numerical values of interaction coefficients are reported only for significant biotic interactions (90% CRI)
Discussion
Here we used hierarchical N-mixture models, accounting for imperfect detection and symmetric species interactions, to model the effect of species interaction on residual abundance, after accounting for differential landscape requirements across a Mediterranean reptile community. We found evidence of reduced species interactions in the studied community, finding a significant correlation only for two out of the 15 species pairs tested. We a priori hypothesised that in case of inter-specific competition between community members from the same trophic level, negative abundance relationships should occur, while predator–prey interactions occurring between different trophic levels could either produce positive or negative abundance correlations (Abrams 2000). Actually, we did not observe significant interactions occurring between community members clearly belonging to two different trophic levels: that is, estimated interaction coefficients between snakes (i.e. predators) and lizards (i.e. prey) were not significant. Instead, a negative significant correlation on residual abundance occurred between the Common wall lizard (P. muralis) and the Italian three-toed skink (C. chalcides). In the study area we estimated significantly different abundances for these species, P. muralis being more abundant that C. chalcides. Moreover, opposite responses to certain landscape features were observed for these species. Both species prey mainly on invertebrates, C. chalcides having a diet more restricted to Araneae (Ciracì et al. 2022), while P. muralis, also including Araneae as a main item of its diet, possesses a wider trophic niche (Scali et al. 2016). In this context, the negative abundance interaction observed between these potentially competing species can be possibly explained by competitive exclusion (MacArthur and Levins 1964), where the more generalist, opportunistic feeder, P. muralis outperforms C. chalcides for food resources. By contrast, we observed a positive local residual abundance correlation between the Green lizard (L. bilineata) and the smaller Italian wall lizard (P. siculus), despite these species showed different responses to landscape features. While negative interactions between potentially competing species are easily explained by competitive exclusion (MacArthur and Levins 1964), positive interactions are more difficult to interpret. These kind of positive abundance correlations can be explained by mutualistic interactions or facilitation among species (e.g. Levi et al. 2013), but none of the two are known to occur among our reptile species so far. Alternatively, positive significant correlation between two potential competitors can arise from the species selecting the same habitat or resource, which has not been accounted for in the model, such as food resources or microclimatic conditions (e.g. Brodie et al. 2018). Both lizards are known to be generalist opportunistic predators, their diet being mainly composed by invertebrate prey (Angelici et al. 1997; Zuffi and Giannelli 2013), despite the larger species (L. bilineata) is known to occasionally include in its diet other lizard species, including P. siculus (Angelici et al. 1997). In this perspective, the observed positive relationship between the two species could also be partially explained as a result of a potential predator–prey interaction: that is, an interplay of both shared selection for unaccounted habitat/resources and an occasional predation of the Green lizard on the Italian wall lizard could exist. Since P. siculus is the numerically dominant species in the community and L. blineata has a markedly lower abundance, occasional predation on P. siculus should not display a strong negative effect on prey density, and in turn could contribute to release both species from inter-specific competition for invertebrate resources (Chase et al. 2002).
Linking biodiversity responses in terms of community composition and abundance to landscape features is of paramount importance for defining management plans and functional conservation programmes (e.g. Lindenmayer and Franklin 2002), however it requires a consistent response of the community to landscape structures (Watson et al. 2005). By contrast, in our study we observed differential responses of species to our set of predictors: nevertheless, some principles can still be drawn. For instance, NPP, when significant for a given species (i.e. L. bilineata, P. muralis, C. chalcides), always had a negative effect on local abundance. This result can be explained because, in the study area, patches with higher NPP are matched to intensive agricultural activities, where mechanized agriculture (e.g. hay meadows), tilling, input of nutrients from livestock manures and pesticides are present, thus determining a disturbance for the reptile community (Rotem et al. 2013; Guiller et al. 2022). The reptile community instead showed a differential effect for other landscape metrics such as TCD: abundance of P. muralis was positively influenced by tree cover, while the congener P. siculus and the skink C. chalcides were negatively affected by this feature. This finding is in line with known ecological requirements and habitat selection preferences of the studied species: in central and southern Italy P. muralis being more frequently associated with woodlands, in particular in shady, humid and structurally complex environments (Corti et al. 2010), while P. siculus and C. chalcides select ecotonal or open areas with lower density of arboreal vegetation (Corti et al. 2010). This is also confirmed by the significant effect of EDGE density on C. chalcides, indicating that local abundance of this species is positively influenced by a higher density of ecotone between woody vegetation and open areas, a landscape structure already recognized as important for herpetofauna across the Mediterranean region (e.g. Atauri and De Lucio 2001; Schindler et al. 2013). Finally, landscape/land-use heterogeneity (LUH) had a significant negative effect on P. muralis. Being this species positively associated to forest cover, this result may indicate that high levels of landscape heterogeneity, at least at the studied spatial resolution, in our context may negatively affect suitability of arboreal vegetated patches for this species requirement.
One of the major criticisms moved to the environmental filtering conceptual framework deals with the difficulty/impossibility of decoupling the effect of the environment from the one of the biotic interactions, at least unless complex experimental designs involving community manipulation are implemented (Kraft et al. 2015; Cadotte and Tucker 2017; Germain et al. 2018). That is, simple observational data are of difficult interpretation in the framework of environmental filtering, because the effect of environment and competition are confounded (Kraft et al. 2015; Germain et al. 2018). These criticisms may partly arise from the lack of an adequate analytical toolbox to treat observational data in the framework of environmental filtering: that is, observed data from a realized community are not only a product of environmental filtering and biotic filtering processes, but are also the realisation of a “detection filter” occurring during sampling, that should be considered (Roth et al. 2018). In this respect, within our reptile community Mean individual detection probability was very low (i.e., p ~ 0.10). Therefore, not properly accounting for this detection error, may result in a biased estimation of local abundances up to 80%, thus producing cascade bias for both the environmental and biotic filtering processes. Moreover, since we also observed differential temporal dynamics in the abundance of our study community, we stress that in addition to the “detection filter”, also the effect of a “temporal dynamic filter” should be accounted for, when deriving inferences from realized communities over consecutive years. As a matter of fact, the application of hierarchical N-mixture models allows to properly address these issues. To date, applications of N-mixture models with symmetric interactions to specifically partition the contribution of environmental and biotic filtering are scarce and restricted to bird communities in forest landscapes, but nevertheless encouraging (Basile et al. 2021; Montano-Centellas et al. 2021). Our results confirmed that the application of these models based on cost-effective count data, accounting for both imperfect detection and temporal dynamics, represent a viable way to disentangle the effect of the environment and biotic interactions on the filtering process of the local abundance of a Mediterranean reptile community. However, despite binomial N-mixture models accounting for species interactions provided reliable results in the study case, some limitations are present and should be considered. Indeed, only 6 out of 12 species from the realized community could have been included in the analysis, given the low number of observations recorded during the sampling. Low detection rates are particularly common for reptiles, representing a severe limitation when the goal is to obtain reliable abundance estimates or fine ecological inferences (Mc Diarmid et al. 2012), binomial N-mixture also being data demanding (Kéry and Royle 2015, 2020). For this reason, although considering that very rare species have a limited ecological relevance on the biotic interaction across the realized community, finding new tools to allow analysis of rare species is of paramount importance. In this respect, multinomial N-mixture models, relying on different sampling protocols, such as multiple observers (Costa et al 2020; Romano et al. 2021) or mark-recapture distance sampling (Rosa et al. 2022b), have proved to be a reliable and cost-effective tool to estimate abundance and other demographic parameters of amphibians and reptile populations, even when density and detection probability are low (Costa et al 2020). Therefore these model may represent a promising pathway for inferring species co-occurrence and interaction also for rare reptile species.
Data availability
The data generated and analysed during the current study are available from the corresponding author on reasonable request.
References
Abrams PA (2000) The evolution of predator-prey interactions: theory and evidence. Ann Rev Ecol Syst 31:79–105
Atauri JA, De Lucio JV (2001) The role of landscape structure in species richness distribution of birds, amphibians, reptiles and lepidopterans in Mediterranean landscapes. Landsc Ecol 16:147–159
Barker RJ, Schofield MR, Link WA, Sauer JR (2018) On the reliability of N-mixture models for count data. Biometrics 74:369–377
Basile M, Asbeck T, Cordeiro Pereira JM, Mikusiński G, Storch I (2021) Species co-occurrence and management intensity modulate habitat preferences of forest birds. BMC Biol 19:1–15
Blondel J (2006) The ‘design’ of Mediterranean landscapes: a millennial story of humans and ecological systems during the historic period. Human Ecol 34:713–729
Blondel J, Aronson J (1999) Biology and wildlife of the Mediterranean region. Oxford University Press, New York
Brodie JF, Helmy OE, Mohd-Azlan J, Granados A, Bernard H, Giordano AJ, Zipkin E (2018) Models for assessing local-scale co-abundance of animal species while accounting for differential detectability and varied responses to the environment. Biotropica 50:5–15
Brotons L, Herrando S, Martin JL (2005) Bird assemblages in forest fragments within Mediterranean mosaics created by wildfires. Landsc Ecol 19:663–675
Burel F, Baudry J (1995) Species biodiversity in changing agricultural landscapes: a case study in the Pays d’Auge, France. Agric Ecosyst Environ 55:193–200
Cadotte MW, Tucker CM (2017) Should Environmental Filtering be Abandoned? Trends Ecol Evol 32:429–437
Casazza G, Malfatti F, Brunetti M, Simonetti V, Mathews AS (2021) Interactions between land use, pathogens, and climate change in the Monte Pisano, Italy 1850–2000. Landsc Ecol 36:601–616
Chase JM, Abrams PA, Grover JP, Diehl S, Chesson P, Holt RD, Case TJ (2002) The interaction between predation and competition: a review and synthesis. Ecol Lett 5:302–315
Ciracì A, Razzetti E, Pavesi M, Pellitteri-Rosa D (2022) Preliminary data on the diet of Chalcides chalcides (Squamata: Scincidae) from Northern Italy. Acta Herpetol 17:71–76
Clare JD, Linden DW, Anderson EM, MacFarland DM (2016) Do the antipredator strategies of shared prey mediate intraguild predation and mesopredator suppression? Ecol Evol 6:3884–3897
Corti C, Capula M, Luiselli L, Razzetti E, Sindaco R (2010) Fauna d’Italia Vol. XLV—Reptilia. Calderini, Bologna, Italy.
Costa A, Romano A, Salvidio S (2020) Reliability of multinomial N-mixture models for estimating abundance of small terrestrial vertebrates. Biodivers Conserv 29:2951–2965
Costa A, Salvidio S, Penner J, Basile M (2021) Time-for-space substitution in N-mixture models for estimating population trends: a simulation-based evaluation. Sci Rep 11:4581
Cox N, Young BE, Bowles P, Fernandez M, Marin J, Rapacciuolo G, Böhm M et al (2022) A global reptile assessment highlights shared conservation needs of tetrapods. Nature 605:285–290
Dormann CF, Elith J, Bacher S, Buchmann C, Carl G, Carré G, García Marquéz R et al (2013) Collinearity: a review of methods to deal with it and a simulation study evaluating their performance. Ecography 36:27–46
Duarte A, Adams MJ, Peterson JT (2018) Fitting N-mixture models to count data with unmodeled heterogeneity: bias, diagnostics, and alternative approaches. Ecol Model 374:51–59
European Environment Agency (2019c) High Resolution Layer: Small Woody Features (SWF) 2015 v. 1.2, released under the framework of the Copernicus programme.
European Environment Agency (2019a) Corine Land Cover (CLC) 2018, Version 2020_20u1, released under the framework of the Copernicus programme.
Farina A (1997) Landscape structure and breeding bird distribution in a sub-Mediterranan ago-ecosystem. Landsc Ecol 12:365–378
Gearty W, Carrillo E, Payne JL (2021) Ecological filtering and exaptation in the evolution of marine snakes. Am Nat 198:506–521
Gelman A, Rubin DB (1992) Inference from iterative simulation using multiple sequences. Stat Sci 7:457–511
Germain RM, Mayfield MM, Gilbert B (2018) The ‘filtering’metaphor revisited: competition and environment jointly structure invasibility and coexistence. Biol Lett 14:20180460
Guiller G, Legentilhomme J, Boissinot A, Blouin-Demers G, Barbraud C, Lourdais O (2022) Response of farmland reptiles to agricultural intensification: Collapse of the common adder Vipera berus and the western green lizard Lacerta bilineata in a hedgerow landscape. Anim Conserv. https://doi.org/10.1111/acv.12790
Jung M (2016) LecoS—A python plugin for automated landscape ecology analysis. Ecol Inform 31:18–21
Keddy PA (1992) Assembly and response rules: two goals for predictive community ecology. J Veg Sci 3:157–164
Kellner K (2015) jagsUI: a wrapper around rjags to streamline JAGS analyses. R package version, 1(1).
Kéry M (2018) Identifiability in N-mixture models: A large-scale screening test with bird data. Ecology 99:281–288
Kéry M, Schaub M (2011) Bayesian population analysis using WinBUGS: a hierarchical perspective. Academic Press, New York
Kéry M, Royle JA (2015) Applied hierarchical modeling in ecology: Analysis of distribution, abundance and species richness in R and BUGS: Volume 1: Static models. Academic Press.
Kéry M, Royle JA (2020) Applied hierarchical modeling in ecology: analysis of distribution, abundance and species richness in R and BUGS: Volume 2: Dynamic and advanced models. Academic Press, London
Kraft NJB, Adler PB, Godoy O, James EC, Fuller S, Levine JM (2015) Community assembly, coexistence and the environmental filtering metaphor. Funct Ecol 29:592–599
Lattanzi E, Perinelli E, Riggio E (2004) Flora vascolare del bosco di Foligno (Nettuno-Roma). Inf Bot Italiano 36:337–361
Levi T, Silvius KM, Oliveira LF, Cummings AR, Fragoso JM (2013) Competition and facilitation in the capuchin–squirrel monkey relationship. Biotropica 45:636–643
Lindenmayer DB, Franklin JF (2002) Conserving forest biodiversity: a comprehensive multiscale approach. Island Press, Washington, DC
Link WA, Schofield MR, Barker RJ, Sauer JR (2018) On the robustness of N-mixture models. Ecology 99:1547–1551
MacArthur R, Levins R (1964) Competition, habitat selection, and character displacement in a patchy environment. Proc Natl Acad Sci USA 51:1207–1210
McDiarmid RW, Foster MS, Guyer C, Chernoff N, Gibbons JW (2012) Reptile biodiversity: standard methods for inventory and monitoring. University of California Press, Berkeley
Montaño-Centellas FA, Loiselle BA, Tingley MW (2021) Ecological drivers of avian community assembly along a tropical elevation gradient. Ecography 44:574–588
Myers N, Mittermeier RA, Mittermeier CG, Da-Fonseca GAB, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403:853–858
Ovaskainen O, Abrego N (2020) Joint species distribution modelling: with applications in R. Cambridge University Press, Cambridge
Plieninger T, Hui C, Gaertner M, Huntsinger L (2014) The impact of land abandonment on species richness and abundance in the Mediterranean Basin: a meta-analysis. PLoS ONE 9:e98355
Plummer M (2003) JAGS: A program for analysis of Bayesian graphical models using Gibbs sampling. In: Proceedings of the 3rd International Workshop on Distributed Statistical Computing, vol 124, pp 1–10
Pollock LJ, Tingley R, Morris WK, Golding N, O’Hara RB, Parris KM, McCarthy MA (2014) Understanding co-occurrence by modelling species simultaneously with a Joint Species Distribution Model (JSDM). Methods Ecol Evol 5:397–406
Potts SG, Petanidou T, Roberts S, O’Toole C, Hulbert A, Willmer P (2006) Plant-pollinator biodiversity and pollination services in a complex Mediterranean landscape. Biol Conserv 129:519–529
European Environment Agency (2019b). High Resolution Layer: Tree Cover Density (TCD) 2018, released under the framework of the Copernicus programme.
Ribeiro R, Santos X, Sillero N, Carretero MA, Llorente GA (2009) Biodiversity and Land uses at a regional scale: Is agriculture the biggest threat for reptile assemblages? Acta Oecol 35:327–334
Ricklefs RE (2004) A comprehensive framework for global patterns in biodiversity. Ecol Lett 7:1–15
Romano A, Roner L, Costa A, Salvidio S, Trenti M, Pedrini P (2021) When no color pattern is available: application of double observer methods to estimate population size of the Alpine salamander. Arc Antarct Alp Res 53:300–308
Rosa G, Salvidio S, Costa A (2022a) European plethodontid salamanders on the forest floor: testing for age-class segregation and habitat selection. J Herpetol 56:27–33
Rosa G, Salvidio S, Trombini E, Costa A (2022b) Estimating density of terrestrial reptiles in forest habitats: The importance of considering availability in distance sampling protocols. Trees For People 7:100184
Rotem G, Ziv Y, Giladi I, Bouskila A (2013) Wheat fields as an ecological trap for reptiles in a semiarid agroecosystem. Biol Conserv 167:349–353
Roth T, Allan E, Pearman PB, Amrhein V (2018) Functional ecology and imperfect detection of species. Methods Ecol Evol 9:917–928
Royle JA (2004) N-mixture models for estimating population size from spatially replicated counts. Biometrics 60:108–115
Ruiz JP (1990) Development of Mediterranean agriculture: an ecological approach. Landsc Urban Plan 18:211–220
Running S, Zhao M (2021) MODIS/Terra Net Primary Production Gap-Filled Yearly L4 Global 500m SIN Grid V061. NASA EOSDIS Land Processes DAAC.
Santos X, Cheylan M (2013) Taxonomic and functional response of a Mediterranean reptile assemblage to a repeated fire regime. Biol Conserv 168:90–98
Scali S, Sacchi R, Mangiacotti M, Pupin F, Gentilli A, Zucchi C, Sannolo M, Pavesi M, Zuffi MA (2016) Does a polymorphic species have a ‘polymorphic’diet? A case study from a lacertid lizard. Biol J Linn Soc Lon 117:492–502
Schindler S, von Wehrden H, Poirazidis K, Wrbka T, Kati V (2013) Multiscale performance of landscape metrics as indicators of species richness of plants, insects and vertebrates. Ecol Indic 31:41–48
Sirami C, Nespoulous A, Cheylan JP, Marty P, Hvenegaard GT, Geniez P, Schatz B, Martin JL (2010) Long-term anthropogenic and ecological dynamics of a Mediterranean landscape: Impacts on multiple taxa. Landsc Urban Plan 96:214–223
Swenson NG, Enquist BJ, Phiter J, Kerkhoff AJ, Boyle B, Weiser MD, Elser JJ et al (2012) The biogeography and filtering of woody plant functional diversity in North and South America. Glob Ecol Biogeog 21:798–808
Tobler MW, Kéry M, Hui FK, Guillera-Arroita G, Knaus P, Sattler T (2019) Joint species distribution models with species correlations and imperfect detection. Ecology 100:e02754
Waddle J, Dorazio RM, Walls S, Rice K, Beauchamp J, Schuman M (2010) A new parameterization for estimating co-occurrence of interacting species. Ecol Appl 20:1467–1475
Watson JE, Whittaker RJ, Freudenberger D (2005) Bird community responses to habitat fragmentation: how consistent are they across landscapes? J Biogeog 32:1353–1370
Whitfeld TJS, Kress WJ, Erickson DL, Weiblen GD (2012) Change in community phylogenetic structure during tropical forest succession: evidence from New Guinea. Ecography 35:821–830
Zuffi MA, Giannelli C (2013) Trophic niche and feeding biology of the Italian wall lizard, Podarcis siculus campestris (De Betta, 1857) along western Mediterranean coast. Acta Herpetol 8:35–39
Acknowledgements
We are grateful to Dino Biancolini and Domenico Cascianelli for their support during field activity. We are grateful to two anonymous Reviewers and to Jochen Krauss for their valuable comments on a previous version of the study.
Funding
Open access funding provided by Università degli Studi di Genova within the CRUI-CARE Agreement. Author A.C. is funded by the Italian National Operative Programme “Research and Innovation” (PON—Ricerca e Innovazione, tematica GREEN; CUP N. D31B21008270007).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Fieldwork and data collection and were performed by AR and RN. Data analyses were performed by AC and GR. The first draft of the manuscript was written by AC, GR and SS and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Romano, A., Rosa, G., Salvidio, S. et al. How landscape and biotic interactions shape a Mediterranean reptile community. Landsc Ecol 37, 2915–2927 (2022). https://doi.org/10.1007/s10980-022-01517-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10980-022-01517-6