Abstract
Since the beginning of the Anthropocene, lacustrine biodiversity has been influenced by climate change and human activities. These factors advance the spread of harmful cyanobacteria in lakes around the world, which affects water quality and impairs the aquatic food chain. In this study, we assessed changes in cyanobacterial community dynamics via sedimentary DNA (sedaDNA) from well-dated lake sediments of Lake Tiefer See, which is part of the Klocksin Lake Chain spanning the last 350 years. Our diversity and community analysis revealed that cyanobacterial communities form clusters according to the presence or absence of varves. Based on distance-based redundancy and variation partitioning analyses (dbRDA and VPA) we identified that intensified lake circulation inferred from vegetation openness reconstructions, δ13C data (a proxy for varve preservation) and total nitrogen content were abiotic factors that significantly explained the variation in the reconstructed cyanobacterial community from Lake Tiefer See sediments. Operational taxonomic units (OTUs) assigned to Microcystis sp. and Aphanizomenon sp. were identified as potential eutrophication-driven taxa of growing importance since circa common era (ca. CE) 1920 till present. This result is corroborated by a cyanobacteria lipid biomarker analysis. Furthermore, we suggest that stronger lake circulation as indicated by non-varved sediments favoured the deposition of the non-photosynthetic cyanobacteria sister clade Sericytochromatia, whereas lake bottom anoxia as indicated by subrecent- and recent varves favoured the Melainabacteria in sediments. Our findings highlight the potential of high-resolution amplicon sequencing in investigating the dynamics of past cyanobacterial communities in lake sediments and show that lake circulation, anoxic conditions, and human-induced eutrophication are main factors explaining variations in the cyanobacteria community in Lake Tiefer See during the last 350 years.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The upsurge in human fertilizer and land-use change during the Anthropocene has intensified nutrient cycling. Further acceleration of nutrient cycling by global climate change has consequently resulted in a change in the biodiversity of natural communities in aquatic ecosystems (Steffen et al. 2007; Taranu et al. 2015; Irfan and Alatawi 2019). Among the most susceptible ecosystems to anthropogenic activities and biodiversity losses is the freshwater ecosystem (Adrian et al. 2009), where cyanobacteria dominance has resulted, amongst others, from intensified land usage and inordinate discharge of nutrients into lakes (Taranu et al. 2015). Perhaps the first direct consequence of cyanobacteria blooms is experienced by diatoms and organisms of higher trophic levels (Dokulil and Teubner 2000), because cyanobacteria can outcompete the eukaryotic phytoplankton due to their rapid growth and eco-physiological adaptation to low light levels (Carey et al. 2012; Rigosi et al. 2014).
Eutrophic water bodies can be dominated by toxin-producing cyanobacteria like Microcystis spp. (Bullerjahn et al. 2016). Some cyanobacteria species like Aphanizomenon are able to fix nitrogen thereby promoting their growth under nitrogen-limited conditions accompanied by high temperatures (Breitbarth et al. 2007). Blooms of cyanobacteria form large clusters and colonies that move vertically in the water column by being able to counteract gas vesicle buoyancy via carbohydrate ballast readjustment (Pfeifer 2012). Taken together, these attributes coupled with their high susceptibility to human-induced changes make cyanobacteria ideal indicator organisms for investigating long term changes in lake ecosystems.
Investigating natural archives such as varved (annually laminated) lake sediments has provided us with a better understanding of how aquatic ecosystem communities have responded to anthropogenic changes of the last centuries (Smol 2009). The DNA preserved in sediment records (sedaDNA) has the potential to reconstruct the dynamics of biological communities and, in particular, those taxa that do not leave a distinct morphological footprint (Capo et al. 2017). Information retrieved from varved sedaDNA archives by amplicon sequencing of DNA fragments, i.e., the analysis of specific genes or taxa on a high phylogenetic resolution, have been useful in reconstructing cyanobacterial community from lakes by providing reliable coverage of the entire cyanobacterial phylum as well as historical information on those cyanobacteria group like the picocyanobacteria that do not form resting cysts. Sedimentary DNA has been useful for instance in revealing distinctive distribution of Synechococcus and Microcystis in water and sediment in Lake Taihu (Ye et al. 2011) as well as in revealing homogenization of cyanobacterial communities in the last century in peri-alpine temperate lakes (Monchamp et al. 2018). Therefore, with the continued advancements in sequencing techniques, sedaDNA is well suited to address knowledge deficits in paleolimnology (Domaizon et al. 2017). Studying the patterns of species diversity across space and time of lake sediments with their seasonally resolved archives opens up the possibility to increase the resolution on past human influences (Brauer and Guilizzoni 2004), and understanding how such a pattern has the potential to influence how ecosystems can be managed and conserved (Monchamp et al. 2016).
The effects of climate change and eutrophication (Paerl and Huisman 2009), as well as that of environmental changes based on sedaDNA since the beginning of industrialization on cyanobacteria, are well documented (Monchamp et al. 2018). However, much of the history of cyanobacteria before this time point coupled with how the effects of both natural environmental change and human agricultural practices affect their dynamics remains unclear. Also, of increasing interest in recent years are the non-photosynthetic lineages that share a common ancestry with cyanobacteria (NCY): Melainabacteria and Sericytochromatia, and more studies are trying to understand their ecology in lake ecosystems (Rienzi et al. 2013; Soo et al. 2017; Monchamp et al. 2019). To fill these knowledge gaps, the present study focuses on reconstructing cyanobacterial dynamics in three lake sediment segments from present time back to the Little Ice Age (CE 1665–2006) using an amplicon sequencing approach, with the aim of understanding changes in their community dynamics during this period as archived in the sediments. We selected Lake Tiefer See within the Klocksin lake chain (Lake TSK) as a study system for several reasons, namely; its detailed record of human impact via land use intensity (agricultural practices) and deforestation leading to vegetation openness (inferred from pollen data) (Theuerkauf et al. 2015), its varved and non-varved sediment segments (Kienel et al. 2017), and finally, its well-established sediment chronology based on varve counting, radiocarbon dating, tephrochronology as well as reconstructed oxic and anoxic conditions in the water column indicated by organic matter δ13C (varve preservation proxy; Dräger et al. 2017, 2019). The relevance of an extended reconstruction of cyanobacteria from lake sediments with a reliable age model is that observed community changes can be accurately related to past natural and/or anthropogenic activities potentially responsible for such changes.
Site description
Lake Tiefer See is part of the Klocksin-Lake-Chain (Fig. 1), a subglacial gully system in a morainic terrain located in the natural park ‘Nossentiner/Schwinzer Heide’ (Kienel et al. 2013, 2017; Dräger et al. 2017, 2019). Today, the lake is connected to Lake Hofsee in the South while the connection to Lake Flacher See in the North has been channelized in a tunnel after the construction of a railway dam between the two lakes (CE 1884–1886). Lake Tiefer See has a surface area of about 0.75 km2, and the catchment area of about 5.5 km2 is dominated by glacial till. Although the catchment is mainly used for agriculture (Theuerkauf et al. 2015), the direct shoreline of the lake is covered by a fringe of trees and there is no anthropogenic infrastructure such as buildings and roads at the lakeshore. The lake has a maximum depth of 62–63 m and no major inflow and outflow. The present-day lake water is mesotrophic and the circulation mode is either mono- or dimictic, depending on the formation of a winter ice cover (Kienel et al. 2013). The study site is characterized by a warm-temperate climate at the transition from oceanic to continental conditions. Mean monthly temperatures range from 0 in January to 17–18 °C in July with maxima up to 30 °C and minima down to − 5 °C. Mean monthly precipitation varies between ~ 40 mm during winter and ~ 60 mm in summer with a mean annual precipitation of 560–570 mm.
a Bathymetric map, b location of Tiefer See in the Klocksin Lake Chain and c land cover in the catchment area. Black stars show former Slavic settlements. Modified after Dräger et al. (2017)
Materials and methods
Core sampling, chronology and total nitrogen estimation
A surface sediment core of length 115 cm was obtained in September 2015 from the deepest point of Lake Tiefer See (62.5 m water depth), using a 90-mm UWITEC piston corer. The core was sealed and stored vertically at 4 °C in a dark room. The core was split longitudinally, photographed, and described. For molecular analyses, one half of the core was sub-sampled at 35 time points for continuous 1 cm intervals from three different sediment types (Fig. ESM1); recent-varved sediments from 9 to 42 cm depth (CE 2006–1924), non-varved from 43 to 82 cm and 88 to 99 cm (ca. CE 1920–1740 and ca. CE 1715–1675) and subrecent-varved segment from 83 to 87 cm (ca. CE 1735–1720). The uppermost 9 cm were excluded from our analyses because of sediment distortion during coring. Molecular analyses were carried out on two subsampled duplicates. The age model was constructed by a multiple dating approach including microscopic varve counting, radiocarbon dating, and the identification of Askja AD 1875 tephra (Wulf et al. 2016; Dräger et al. 2017).
Total nitrogen (TN) of freeze-dried sediments was measured continuously at 1-cm increments from bulk samples with an elemental analyzer (EA 3000-CHNS Eurovector) as previously described in Dräger et al. (2017).
Nucleic acids extraction
The top contaminated layers were scraped off with a sterile razor before the uncontaminated anoxic part was put into sterile 15-ml falcon tubes and stored at − 80 °C until analysis. Care was taken to prevent cross contamination of the sediments by processing recent and older samples at different times.
The sediment samples were homogenized following the mortar method described by Herrera and Cockell (2007). All DNA extraction equipment was placed under a UV lamp in a laminar flow hood for a minimum of 25 min before use. DNA was then extracted from approximately 300 mg of sediment sample in duplicates, using the PowerSoil DNA Isolation Kit (Qiagen). Briefly, following initial chemical and mechanical lysis steps where DNA is separated from debris via centrifugation, proteins are precipitated and then the DNA is purified with silica spin filters. DNA extractions were done in batches of seven samples with the addition of one negative control each time while avoiding contamination from foreign DNA by following processes described in Monchamp et al. in 2016. Extracted sedaDNA was stored at − 20 °C until further downstream analyses.
DNA preparation and amplification
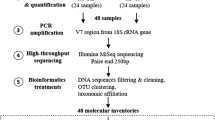
Library preparation for Illumina high-throughput sequencing (HTS) was performed via polymerase chain reaction (PCR) using previously published cyanobacterium-specific primers CYA106F (5′ CGGACGGGTGAGTAACGCGTG 3′) (Nübel et al. 1997), and CYA784R (5′ ACTACWGGGGTATCTAATCCC 3′) (Monchamp et al. 2016). These amplify a 678-nt-long fragment of the V1–V4 regions of the 16S rRNA gene. Added to the primers were overhanging barcodes for sample differentiation. Being the first study of its kind in this lake, it was imperative that the primer set covers most taxa all over the cyanobacteria phylum. This phylum coverage was tested in silico against the ARB-SILVA database v128 (Klindworth et al. 2013).
SedaDNA samples including positive control (Microcystis aeruginosa PCC7806) and negative isolation control were amplified in a PCR of 50 µl, containing 10 × Pol Buffer C (Roboklon GmbH, Berlin, Germany), 25 mM MgCl, 0.2 mM deoxynucleoside triphosphate (dNTP) mix (ThermoFisher Scientific, Massachusetts, USA), 0.5 mM each primer (TIB Molbiol, Berlin, Germany) and 1.25 U of Optitaq Polymerase (Roboklon). The volume of the template DNA used in each reaction varied between 1 and 4 µl depending on the sedaDNA concentration. The thermal cycler PCR program included a first denaturation step at 95 °C for 10 min, followed by 35 cycles at 95 °C for 15 s, annealing at 60 °C for 30 s, extension at 72 °C for 45 s and a final extension step at 72 °C for 5 min. PCR products were purified with Agencourt AMPure XP kit (Beckman Coulter, Nyon, Switzerland) and eluted in 30 µl DNA/RNA-free water. The purified product was quantified with a Qubit (2.0) fluorometer. The unique barcoded samples together with the positive and negative controls were pooled in equimolar concentration into one library. The final library was sent to Eurofins Scientific (Constance, Germany) for sequencing using the Illumina MiSeq v3 kit (2 × 300 bp).
Bioinformatics and statistical analyses
Due to the long amplicon lengths, the forward (R1) and reverse reads (R2) were not merged during processing but treated separately as R1 and R2. From 35 samples a total of 5,463,680 reads (R1) and 687,801 reads (R2) were obtained. The raw reads were quality checked with FastQC (Andrews et al. 2015). Raw data were demultiplexed using Cutadapt (Martin 2014). Primer sequences were allowed to have 10% mismatches. Phred-Scores of the barcodes were required to be higher than Q35 and no mismatches were allowed during sorting. Sequences were further processed using Trimmomatic to trim and filter out low-quality sequences (Bolger et al. 2014). USearch (Edgar 2010) (v6.) was used to identify and remove chimeric sequences. After quality-filtering, chimera removal and primer trimming 4,505,289 (R1) and 570,095 (R2) reads remained. The negative control had one read in R1 and zero reads in R2 and was therefore removed. The script pick_open_reference.py from the Quantitative Insights into Microbial Ecology (QIIME) pipeline was used for Operational Taxonomic Unit clustering (OTU) with a minimum nucleotide sequence similarity of 97% (Caporaso et al. 2010) and SILVA database (v128) for taxonomic assignment (Quast et al. 2013).
Before statistical analyses, singletons, OTUs assigned to other bacteria, archaea, chloroplasts, and mitochondria were removed after which 1,134,146 (R1) and 205,775 (R2) cyanobacteria reads remained. The positive control had 250 (R1) and 58 (R2) reads with the OTUs assigned only to Microcystis spp., which did not occur in our samples thus ruling out cross-contamination and was consequently removed. OTUs with less than 0.5% of total read counts and the unclassified cyanobacteria OTUs were filtered out. Cyanobacteria absolute read counts were Hellinger-transformed prior to beta diversity analyses (Legendre and Gallagher 2001). We used principal coordinates analysis (PCoA) with Bray–Curtis distance to visualize distances among the investigated sediment segments from R1 and R2. The variance explained by the PCoA axes from R1 and R2 were comparable (ESM2A, B). Based on this observation and the low number of reads recovered from the R2 data, we therefore carried out further statistical analyses using only the R1 data. For alpha diversity and richness estimation, we compared both rarefied [using the rtk package in R (Saary et al. 2017)] and unrarefied absolute OTU read counts and found them to be identical (ESM3), thus, we present and discuss data only from the unrarefied read counts. Variation in cyanobacterial OTU composition, diversity among the samples, t tests for each non-photosynthetic cyanobacteria sister clade separated by varved/non-varved periods, and the different time segments (recent-varved, non-varved and subrecent-varved sediments), as well as relationships with environmental parameters, were determined with PAST3.14 (Hammer et al. 2001), and CANOCO5 (Smilauer and Leps 2014). A non-parametric permutational multivariate analysis of variance (PerMANOVA) based on the Bray–Curtis dissimilarity index was used to test for significant differences in the overall cyanobacterial communities among the three time segments. Due to a low number of reads, five samples from the deep non-varved segment ca. CE 1715–1675 (88–99 cm) were removed before ordination analyses. Environmental data from the different sediment segments were standardized by subtracting the mean and dividing by the standard deviation prior to analyzing their variation in a principal component analysis (PCAs) based on Euclidean distance. Environmental data was then fitted to a distance-based Redundancy Analysis (dbRDA; Bray–Curtis distance) model as explanatory variables with cyanobacteria community data as response variables using the “capscale” function in the vegan package in R (Oksanen et al. 2019). The overall significance of the dbRDA model and its axes were tested by a 999 Monte Carlo permutation test using the “anova.cca” function in vegan. The subset of explanatory variables that contributed significantly to the dbRDA model was further tested by forward selection permutation analysis (999 permutations with pseudo-F statistical test) using the “ordiR2step” function in vegan. Canonical variation partitioning analysis (VPA) (Legendre et al. 2005; Ramette and Tiedje 2007) with redundancy analysis (RDA) was implemented on three categories of environmental variables, that is, eutrophication (TN), lake circulation (reconstructed vegetation openness and δ13C) and other factors (dry density and CaCO3) to identify their unique and combined effects on cyanobacteria community structure. The significance of the fractions of variability explained by the categories was tested and determined via 999 Monte Carlo permutation in CANOCO5. Trend analysis of absolute read count changes of non-photosynthetic cyanobacteria sister clades during the period investigated was done with Microsoft Excel.
Sequencing data and metadata are available at the European Nucleotide Archive (ENA) under BioProject accession number PRJEB33742 and sample accession numbers ERS3606196-ERS3606230.
Phylogenetic analyses of the OTUs
To verify the accuracy of the taxonomic assignment based on SILVA, we used the OTU FASTA sequences from Lake Tiefer See to construct a cyanobacteria phylogenetic tree according to Komárek et al. (2014). Sequences were aligned in the arb software (Ludwig et al. 2004) using the integrated sequence alignment tool under the default setting. Acidithiobacillus thiooxidans strain ATCC 19,377 was added as an outgroup, and 10 cyanobacteria reference sequences from GenBank and ARB-SILVA database (Quast et al. 2013) were also added. The alignment was used to build a bootstrap tree with 100,000 trees based on neighbour-joining. The online tool iTOL was used to visualize and annotate the tree (Letunic and Bork 2016).
Lipid biomarker analysis
6Me-C17:0 and 7Me-C17:0 monomethyl branched alkanes (6- and 7-methylheptadecane) are produced mainly by heterocystous, ramified and some filamentous cyanobacteria (Liu et al. 2013; Coates et al. 2014; Bauersachs et al. 2017) and might be representative of cyanobacterial biomass as recently shown for the Baltic Sea (Kaiser et al. 2020). Sediment samples (n = 5) from a parallel core correlated to the reference core based on varve counting (not shown) were analyzed following (Kaiser et al. 2016, 2020). Sediments (0.4 g dry weight) were extracted with a mixture of DCM/MeOH (9:1) and accelerated solvent extraction (Dionex ASE 350). Squalane was added as an internal standard. The apolar lipid fractions containing the hydrocarbons were obtained by column chromatography (SiO2) using hexane as eluent. The fractions were analyzed by gas chromatography–mass spectrometry (GC–MS) using an Agilent Technologies 7890B GC system coupled to a 5977B Mass Selective Detector equipped with an HP-5 ms capillary column (30 m * 0.25 mm * 0.25 µm). The oven temperature was programmed from 40 to 320 °C at 8 °C min−1 followed by a 15 min isotherm. GC–MS data were collected in total ion current (TIC) (m/z 50–600). The fractions were also analyzed using a multichannel TraceUltra gas chromatograph (ThermoScientific) equipped with a DB-5 ms capillary column (30 m * 0.25 mm * 0.25 µm) and a flame ionization detector (FID). The temperature program was identical to that used for analysis by GC–MS and peak identification was made by comparison of retention times using the two methods. For quantification, the GC-FID response of each lipid was normalized to that of the internal standard and the amount of sediment extracted. The detection limit was estimated to ca. 4–5 ng g−1. The data were further normalized to total organic carbon (TOC).
Results
Cyanobacterial communities reconstructed from Lake Tiefer See sediments
A total of 146 OTUs spanning the entire cyanobacteria phylum was obtained from 35 sedaDNA samples (from ca. CE 1670–2006; Fig. 2). Almost all the OTUs were assigned to photosynthetic cyanobacteria 89% (130 OTUs) and 5.5% were unclassified (8 OTUs). Of the 130 OTUs assigned to photosynthetic cyanobacteria, 120 could be assigned to the order level, 111 to the family level, and 90 were assigned to the genus level. No OTU was assigned to a species level with high confidence (confidence threshold 0.95).
Phylogenetic tree constructed from all the 145 OTUs reference sequences from the sediment samples of Lake Tiefer See based on the neighbour-joining method. Acidothiobacillus thiooxidans was used as root with the addition of 10 cyanobacteria reference sequences sourced from the GenBank and ARB-SILVA database. The colors represent the different cyanobacteria orders. The 20 most abundant OTUs are marked with a star, those marked with the ‘^’ symbol are the representative sequences and the reference sequences are in bold fonts
Richness and diversity
Our study showed that sedaDNA was preserved and concentrations declined from CE 1920 downcore (~ 40 cm depth) with richness and diversity indices showing a similar trend (Fig. 3). High throughput sequencing data revealed cyanobacteria richness to be highest in recent-varved sediments for the period CE 2006–1924 (9–42 cm depth) and ranged between 69 and 14 (mean richness was 35). Likewise, richness in subrecent-varved sediments for the period ca. CE 1735–1720 (83–87 cm) was higher and ranged between 21 and 27 (mean richness was 24). In contrast, richness was lower in non-varved sediments for the period ca. CE 1920–1740 (43–82 cm) compared to both varved sediment segments and ranged between 10 and 21 (mean 16). Similarly, the Shannon diversity index ranged between 2.5–3.3 (mean 2.8) and 2.7–2.8 (mean 2.8) in recent- and subrecent-varved sediments, respectively. Lower diversity was recorded in the non-varved sediments and ranged from 2.0 to 2.7 (mean 2.5). Furthermore, a one-way non-parametric permutational multivariate analysis of variance (PerMANOVA) revealed a significant variation in the total cyanobacteria community composition among the three time segments (P = 0.0001, F = 5.1; ESM4).
Spatio-temporal variation of sediment characteristics, molecular and geochemical analyses. From left to right: log10 transformed sedimentary DNA concentration (sedaDNA), Shannon index (diversity), Richness (no. of OTUs). The TOC content, reconstructed vegetation openness, TN, and δ13C were published in Dräger et al. (2017, 2019). Dark grey-shaded and non-shaded areas reflect periods of weakened and intensified lake circulation, respectively
Most abundant OTUs
Twenty OTUs predominated the sediments of Lake Tiefer See in the period investigated (1678–2006; Fig. 4). Out of the total 1,134,146 cyanobacteria read counts, 81.5% (924,329 reads) were assigned to the 20 most abundant OTUs. The remaining 126 OTUs amounted to 18.5% (209,817 reads). The most abundant OTUs were assigned to the picocyanobacteria Cyanobium PCC-6307 belonging to the order Synechococcales, and the heterocystous Aphanizomenon NIES81 belonging to the order Nostocales.
Spatio-temporal variation of the relative abundance of the 20 most abundant OTUs. The SILVA database (version 128) was utilized for open-reference OTU clustering (97% sequence similarity) and taxonomic assignments. Dark grey-shaded and non-shaded areas reflect periods of weakened and intensified lake circulation, respectively
Non-photosynthetic cyanobacteria sister clades
From all OTUs, 5.5% were assigned to the non-photosynthetic cyanobacteria (NCY) sister lineages the fermentative Melainabacteria and Sericytochromatia (ML635J-21; 8 OTUs in total; Fig. 5a). Their occurrence spanned the period CE 1990–ca. CE 1720 (n = 12 samples; 14–87 cm; ESM3). Melainabacteria represented by 5 OTUs, had higher richness than the Sericytochromatia with 3 OTUs, however, the latter had more sequencing reads than the former. In the Melainabacteria, most OTUs were assigned to the order Gastranaerophilales (4 OTUs) and 1 OTU to Obscuribacterales (Fig. 5a). Furthermore, the Sericytochromatia group recorded more reads in the non-varved sediments than the varved sediments (n = 8 samples; p = 0.01; Fig. 5b), whereas the Melainabacteria group had significantly more absolute reads in the varved than the non-varved sediments (n = 6 samples; p = 0.001; Fig. 5c). Overall, Shannon diversity for the non-photosynthetic cyanobacteria alone was estimated in the 6 samples with more than 1 OTU (ESM3) and was found, like the richness, to be higher in varved (0.7–1.0) than in non-varved sediments (0.3–0.5; Fig. 5d).
Dynamics of non-photosynthetic cyanobacteria (NCY) sister clades observed in Lake Tiefer See. Absolute reads per (A) OTU of the deep branching Sericytochromatia (ML635J-21) (blue) and Melainabacteria orders Gastraerophilaes (green) and the Obscuribacterales (red). Box plots showing OTU reads of (B) Sericytochromatia (ML635J-21) and (C) Melainabacteria across varved and non-varved sediment segments. (D) Alpha diversity indices (richness and Shannon diversity) of all non-photosynthetic clades over the period investigated. Shaded area in D represents varved period, unshaded area is non-varved period
Cyanobacterial community composition in relation to changes in abiotic conditions
Proxies for environmental variables such as reconstructed vegetation openness for lake mixing and circulation, TN for nutrient availability in the lake, and δ13C for varve preservation in the Lake Tiefer See sediments have been previously published (Dräger et al. 2017, 2019). All environmental data were subject to a principal component analysis (PCAs) to explore variation among the sediment segments; this revealed three distinct, time constrained clusters that contained samples from consecutive depths. The recent-varved sediments CE 2006–1924 (9–42 cm) were characterized by increased TN, TOC, and CaCO3 whereas the non-varved sediments ca. CE 1920–1740 (43–82 cm) and the subrecent-varved sediments CE 1735–1720 (83–87 cm) were defined by elevated values of vegetation openness and δ13C, respectively (ESM5). Furthermore, a distance-based redundancy analysis (dbRDA) was used to determine the relationship between cyanobacterial communities from the sediment segments and environmental variables. A forward selection permutation analysis revealed that TN and CaCO3 best explained the cluster of cyanobacterial OTUs from the recent-varved sediments CE 2006–1924 (9–21 cm), whereas reconstructed vegetation openness and δ13C best explained the cluster in the non-varved segments ca. CE 1920–1740 (43–82 cm) and the subrecent-varved segments ca. CE 1735–1720 (83–87 cm), respectively (all p values < 0.05; Fig. 6a; ESM6). As determined by variation partitioning analysis (VPA), eutrophication (represented by TN; 11.9%) and lake circulation (represented by vegetation openness and δ13C; 16.8%) were the groups of factors that significantly explained most of the variability in cyanobacteria composition in Lake Tiefer See, whereas the interaction between them only accounted for 0.6% and a large part of the variation remained unexplained (that is, 71%; Fig. 6b; ESM7). Other abiotic factors (that is, dry density and CaCO3) when included in the VPA accounted for an additional 6% and increased the total explained variation to 35.2% (ESM8 and 9).
A Distance-based redundancy analysis (dbRDA) based on Bray–Curtis dissimilarity on the effect of environmental variables on cyanobacteria community composition. The dbRDA axes 1 and 2 together explain 35.4% of the variation, while total variation explained by the dbRDA model was 48.4%. The samples from the different sediment segments are color-coded as follows; green = recent-varved, blue = subrecent-varved and red = non-varved. RecVegOp = reconstructed vegetation openness. The environmental variables explaining the variation in cyanobacteria community were all significant (adjusted R2 = 0.38) and are projected as red vectors. The response variables are shown at the genus level. B Venn diagram displaying the results of variation partitioning analysis (VPA). The two explanatory matrices used contained variables about lake circulation and eutrophication. Each circle represents the portion of variation accounted for by each explanatory matrix or a combination of the explanatory matrices. The bounding rectangle represents the total variation in the response data showing the portion of residual variation not explained by the explanatory matrices
Cyanobacterial lipid biomarker reconstructed from sediments
Since the lipid biomarkers 6Me-C17:0 and 7Me-C17:0 monomethyl branched alkanes representative for cyanobacteria elute very close to each other on a DB-5 ms capillary column, their total sum (6 + 7Me-C17:0) was considered hereafter. The TOC normalized content of 6 + 7Me-C17:0 ranged between ~ 5 and 13 µg g−1 TOC. A maximum content occurred in 1973 (13.3 µg g−1 TOC) that correlated with the highest relative abundance of OTUs assigned to Aphanizomenon NIES81 (Fig. 7).
Variability of Aphanizomenon NIES81 and cyanobacteria lipid biomarkers. Abundance of OTUs assigned to Aphanizomenon NIES81 relative to total cyanobacterial OTUs (in %) based on amplicon sequencing compared to 6 + 7Me-C17:0 methylheptadecane lipid biomarker. Data is shown for selected time points since the onset of increasing eutrophication in Lake Tiefer See (since 1924)
Discussion
With our molecular approach, we successfully extracted and sequenced cyanobacterial sedaDNA and reconstructed the cyanobacterial dynamics in Lake Tiefer See sediments spanning the last 350 years. This was made possible due to lake characteristics such as hypoxic to anoxic conditions at the water–sediment interface, the presence of varves, and the absence of burrowing organisms in the sediments which are all conditions known to favor sedaDNA preservation (Coolen et al. 2006; Corinaldesi et al. 2011). We were able to amplify an over 500-nt-long fragment of the cyanobacterial 16S rRNA gene, with a representative coverage of its entire phylum from all samples without enhancements similar to other temperate lakes (Monchamp et al. 2016, 2018; Domaizon et al. 2013). An improved and extended cyanobacteria database (Ramos et al. 2017) was crucial for the phylogenetic analysis of the resulting OTU reference sequences (Fig. 2).
Anthropogenic influence in Lake Tiefer See was so far shown via pollen-based vegetation openness reconstruction (Theuerkauf et al. 2015; Dräger et al. 2017). Anthropogenic deforestation within the catchment during settlement periods probably shaped cyanobacteria dynamics in Lake Tiefer See through the vegetation influence on lake circulation (Dräger et al. 2017). During times of predominant open vegetation, the lake was exposed to stronger wind stress leading to intensified lake circulation and more oxic conditions at the lake bottom which together resulted in the absence of varves (Dräger et al. 2017). Our data revealed a lower diversity in cyanobacteria community composition and a high abundance of OTUs assigned to Cyanobium spp. in non-varved sediments (Fig. 4). The high abundance of Cyanobium concurs with known trends in the water column of temperate lakes where Cyanobium together with other picoplankton cyanobacteria (< 0.2 µM in size) make up the most abundant cyanobacteria (Callieri and Stockner 2000). However, the lower cyanobacteria diversity in non-varved sediments observed herein may be explained by two processes. The increased lake circulation will lead to the disruption of bloom formation by filamentous cyanobacteria species in the water column due to a high water turnover (Visser et al. 2016). When intensified circulation is coupled with nutrient availability, picocyanobacterial groups are favored over large and buoyant taxa. While on the one hand, an increased water turnover may favor sediment deposition of filamentous cyanobacteria, the DNA preservation of these species at the oxygenated water–sediment interface in this freshwater system will be poor (Domaizon et al. 2017). Therefore, reconstructing filamentous cyanobacteria species DNA in high numbers from non-varved sediments which was shown here for Cyanobium spp. becomes less likely. On the other hand, a weaker lake circulation and the resulting anoxic lake bottom favors varve preservation (Kienel et al. 2013, 2017). Constantly lower temperatures at the anoxic lake bottom additionally promote DNA preservation (Coolen et al. 2006). Hence, our results support the hypothesis that the cluster of cyanobacterial OTUs from ca. CE 1720–1735 can be explained by the same conditions that result in varve preservation. An anoxic lake bottom favors DNA preservation and decreased water circulation influences the cyanobacterial community composition (Figs. 3, 6a).
The relationship between anoxic/oxic conditions is further evident through the different distribution of Sericytochromatia (ML536J-21) and the fermentative Melainabacteria in sediments. The OTUs assigned to Sericytochromatia preferably occur in non-varved sediments while those assigned to the Melainabacteria rather occur in varved sediments (P < 0.05; Fig. 5b, c). It has been suggested that members of Sericytochromatia can inhabit both photic and aphotic environments, and since they possess an array of respiratory proteins, they also have members predicted to be capable of aerobic respiration. These members can carry out aerobic respiration under high and low oxygen conditions due to the presence of A- and C-family oxygen reductases in their respiratory mechanisms (Soo et al. 2017). The Melainabacteria have been predicted to adapt to low oxygen and anoxic zones because they possess C-family oxygen reductase and cytochrome bd oxidases (i.e. terminal oxidases that reduces dioxygen to water to avoid the production of reactive oxygen species) in their genomes (Di Rienzi et al. 2013; Soo et al. 2014, 2017). Since lack of knowledge of their metabolism so far hindered the laboratory growth of the non-photosynthetic sister clades of cyanobacteria and thus also research on their physiology (Wrighton et al. 2014), the current knowledge of their metabolism is so far gained largely from culture-independent metagenome reconstructions from environmental samples (Di Rienzi et al. 2013; Soo et al. 2014, 2017). Nevertheless, our observations showing a distinct separation of the NCY from the photosynthetic cyanobacteria clades as well as no increasing trend in their richness over the last three centuries covered in this study concur with similar findings from other temperate lake ecosystems (Monchamp et al. 2019). However, the relationship between oxic/anoxic conditions and the dynamics of the NCY clades which we show here for the first time, emphasizes the need for investigations over longer time scales that encompass more intervals of variations in oxic/anoxic conditions at the lake bottom. Furthermore, since Melainabacteria and Sericytochromatia have been predicted to potentially inhabit a variety of environments like photic and aphotic habitats (Soo et al. 2014, 2017), an in-depth spatio-temporal survey of the water column of Lake Tiefer See would be crucial for evidencing their occurrence in this ecosystem, thus, providing more clarity about their existence.
It has been further shown that agricultural practices in the catchment influenced Lake Tiefer See (Kienel et al. 2013, 2017). Farming in the catchment became intensified with extensive fertilizer application and enhanced transportation of agricultural products after a railway track construction between ca. CE 1884 and 1886. It peaked in the 1970s when pig farming reached 15,000 animals with the resulting manure ending up in the lake (Kienel et al. 2017). Typically, runoffs from agricultural wastes ending up in lakes carry nutrients like nitrogen and phosphorus beneficial to cyanobacteria, in worst cases tipping the scale in favor of eutrophication-associated and potential toxin producing taxa like Aphanizomenon and Microcystis (Preußel et al. 2006; Sukenik et al. 2012). Our reconstruction data show that OTUs assigned to Microcystis and Aphanizomenon, after only a brief occurrence in the early eighteenth century, reappear about CE 1920 (Aphanizomenon) and CE 1977 (Microcystis) and remained part of the lake ecosystem since then (Fig. 4). The permanent occurrence of these potential toxins producing taxa is likely a delayed consequence of the intensification of farming within Lake Tiefer See catchment since ca. CE 1880 with the introduction of mechanized farming, mineral fertilizers (organic human and animal manure) and livestock farming (Kienel et al. 2013). This premise is further supported statistically by dbRDA and VPA (Fig. 6a, b; ESM6), which show that TN was the best predictor for the cluster of OTUs assigned to Microcystis and Aphanizomenon and alone explains 11.9% (VPA) of the variability in cyanobacterial community composition. We therefore suggest that anthropogenic farming practices at least partly caused the observed cyanobacteria dynamics reconstructed from recent-varved sediments in Lake Tiefer See. Increased TN values in the sediments (Fig. 3) were also related to an increase in livestock farming and field drainage in the catchment (Kienel et al. 2013, 2017). Our data revealed significant differences in cyanobacteria community composition between the subrecent (ca. CE 1720–1735) and recent (CE 1924–2005) varved sediments (P < 0.05; ESM4). We therefore further posit that while anoxic conditions are partly the reason for higher diversity in subrecent and recent varved sediments, additional intensive farming led to a significantly stronger change in the temporal variation of cyanobacteria communities in recent varved sediments. From CE 1990, following privatization, livestock farming within the catchment of Lake Tiefer See strongly reduced in line with restoration efforts according to EU agricultural policies resulting in decreased nutrient input into the lake (Theuerkauf et al. 2015; Kienel et al. 2017). Studies have shown that eutrophied lake ecosystems tend to recover when subjected to restoration processes (Jeppesen et al. 2005). However, re-establishing the microbial assemblages that existed pre-fertilization is rarely attained (Bennion et al. 2011). This is what we observe here, namely, the persistence of eutrophication associated cyanobacterial OTUs in Lake Tiefer See almost three decades after human farming practices reduced. Our results therefore suggest that cyanobacterial sedaDNA serves as a potential proxy for lake eutrophication in the past.
Taken together, oxic/anoxic conditions and human-induced eutrophication are important factors driving cyanobacteria dynamics (abundance and community structure) in Lake Tiefer See although most of the variation in cyanobacteria community structure from our study remains unexplained (Fig. 6b). While eutrophication influences the abundance and diversity of the water column cyanobacteria communities, anoxic conditions in the sediment determine how much of the previous communities gets preserved (Domaizon et al. 2017). This means that the sedaDNA archive deposited under oxic conditions most likely does not reflect the entire spectrum of previous water column cyanobacteria communities. Therefore, cyanobacterial sedaDNA is influenced by both the abundance as well as diversity at a given time in the past and by the preservation conditions in the sediment. Furthermore, our study shows that it is difficult to disentangle between both factors in recent varves because eutrophication in part contributes to anoxia in freshwater systems (Schertzer and Sawchuk 1990). The occurrence of higher cyanobacteria diversity in varved sediments at times without intensive agriculture points to the preservation of cyanobacteria sedaDNA as a crucial factor in sediments.
Our sedaDNA data is further supported by lipid biomarkers. In detail, 6- and 7-methylheptadecane are considered as cyanobacterial specific biomarkers predominantly observed in clades including heterocystous, ramified, and some filamentous cyanobacteria, and rarely in unicellular cyanobacteria (Liu et al. 2013; Coates et al. 2014). Since Microcystaceae hydrocarbon branched alkanes profile are absent (Liu et al. 2013), it is very likely not the source organism of 6 + 7Me-C17:0. The hydrocarbon profile of Cyanobium sp. has not been published to the best of our knowledge, but this unicellular picoplankton cyanobacterium most likely does not produce branched alkanes. Recently, Bauersachs et al. (2017) published the hydrocarbon composition of one strain of the genus Aphanizomenon isolated from Baltic Seawater. This strain indeed contained 7Me-C17:0, but in small relative amount (ca. 3%). However, it is possible that other lacustrine strains produce relatively more 7Me-C17:0, and maybe 6Me-C17:0 as well. As records of the TOC normalized content of 6 + 7Me-C17:0 and the relative abundance of OTUs assigned to Aphanizomenon present a similar variability over time (Fig. 7), we suggest that they may quantitatively reflect the presence of filamentous cyanobacteria such as Aphanizomenon sp. in Lake Tiefer See.
Conclusions
Our study provides the first insights into the community structures of cyanobacteria and their non-photosynthetic sister lineages in Lake Tiefer See as archived in sediments that span the last 350 years. We show that lake circulation, anoxic conditions as indicated by varve presence and human-induced eutrophication are the main factors explaining variations in the cyanobacterial community in Lake Tiefer See. A relationship between environmental change and cyanobacterial dynamics (e.g. Sericytochromatia and Melainabacteria) is suggested, thus adding to the cohort of studies making a strong argument for the use of cyanobacterial sedaDNA as a paleo proxy. More importantly, the results from this study add to the growing list of lacustrine ecosystem investigations shedding light on anthropogenic impact on cyanobacterial diversity changes across time and space. We show that eutrophication-associated OTUs were introduced into the lake system mostly as a result of anthropogenic agricultural practices and have since persisted despite restoration efforts. Future investigations will need to focus on exploring the parallels in the dynamics between current water column cyanobacterial communities and those of the sedimentary archive. In addition, reconstructing longer time scales that additionally investigate periods without human influences needs to be done to decipher the role of natural climate variability from anthropogenic effects on cyanobacterial communities.
References
Adrian R, O’Reilly CM, Zagarese H, Baines SB, Hessen DO, Keller W, Livingstone DM, Sommaruga R, Straile D, Donk EV, Weyhenmeyer GA, Winder M (2009) Lakes as sentinels of climate change. Limnol Oceanogr 54:2283–2297. https://doi.org/10.4319/lo.2009.54.6_part_2.2283
Andrews S, Krueger F, Seconds-Pichon A, Biggins F, Wingett S (2015) FastQC. A quality control tool for high throughput sequence data. Babraham Bioinform. Babraham Inst
Bauersachs T, Talbot HM, Sidgwick F, Sivonen K, Schwark L (2017) Lipid biomarker signatures as tracers for harmful cyanobacterial blooms in the Baltic Sea. PLoS ONE 12(10):e0186360. https://doi.org/10.1371/journal.pone.0186360
Bennion H, Battarbee RW, Sayer CD, Simpson GL, Davidson TA (2011) Defining reference conditions and restoration targets for lake ecosystems using palaeolimnology: a synthesis. J Paleolimnol 45:533–544. https://doi.org/10.1007/s10933-010-9419-3
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Brauer A, Guilizzoni P (2004) The record of human/climate interactions in lake sediments. Quat Int 113(1):1–3. https://doi.org/10.1016/s1040-6182(03)00076-4
Breitbarth E, Oschlies A, LaRoche J (2007) Physiological constraints on the global distribution of Trichodesmium: effect of temperature on diazotrophy. Biogeosciences 4:53–61. https://doi.org/10.5194/bg-4-53-2007
Bullerjahn GS, McKay RM, Davis TW, Baker DB, Boyer GL, D’Anglada LV, Doucette GJ, Ho JC, Irwin EG, Kling CL, Kudela RM, Kurmayer R, Michalak AM, Ortiz JD, Otten TG, Paerl HW, Qin B, Sohngen BL, Stumpf RP, Visser PM, Wilhelm SW (2016) Global solutions to regional problems: collecting global expertise to address the problem of harmful cyanobacterial blooms. A Lake Erie case study. Harmful Algae 54:223–238. https://doi.org/10.1016/j.hal.2016.01.003
Callieri C, Stockner J (2000) Picocyanobacteria success in oligotrophic lakes: fact or fiction? J Limnol 59:72–76. https://doi.org/10.4081/jlimnol2000.72
Capo E, Domaizon I, Maier D, Debroas D, Bigler C (2017) To what extent is the DNA of microbial eukaryotes modified during burying into lake sediments? A repeat-coring approach on annually laminated sediments. J Paleolimnol 58(4):1–17. https://doi.org/10.1007/s10933-017-0005-9
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Buschman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widman J, Yatsunenko T, Zanewald J, Knight R (2010) QIIME allows high throughput community sequencing data. Nat Methods 7:335–336. https://doi.org/10.1038/nmeth0510-335
Carey CC, Ibelings BW, Hoffmann EP, Hamilton DP, Brookes JD (2012) Eco-physiological adaptations that favour freshwater cyanobacteria in a changing climate. Water Res 46:1394–1407. https://doi.org/10.1016/j.watres.2011.12.016
Coates RC, Podell S, Korobeynikov A, Lapidus A, Pevzner P, Sherman DH, Allen EE, Gerwick L, Gerwick WH (2014) Characterization of cyanobacterial hydrocarbon composition and distribution of biosynthetic pathways. PLoS ONE 9(1):e85140. https://doi.org/10.1371/journal.pone.0085140
Coolen MJL, Boere A, Abbas B, Baas M, Wakeham SG, Sinninghe Damste JS (2006) Ancient DNA derived from alkenone-biosynthesizing haptophytes and other algae in Holocene sediments from the Black Sea. Paleoceanography 26:PA2204. https://doi.org/10.1029/2005PA001188
Corinaldesi C, Barucca M, Luna GM, Dell’Anno A (2011) Preservation, origin and genetic imprint of extracellular DNA in permanently anoxic deep-sea sediments. Mol Ecol 20(3):642–654. https://doi.org/10.1111/j.1365-294X.2010.04958.x
Di Rienzi SC, Sharon I, Wrighton KC, Koren O, Hug LA, Thomas BC, Goodrich JK, Bell JT, Spector TD, Banfield JF, Ley RE (2013) The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to Cyanobacteria. Elife 2:e01102. https://doi.org/10.7554/eLife.01102.001
Dokulil MT, Teubner K (2000) Cyanobacterial dominance in lakes. Hydrobiologia 698:29–46. https://doi.org/10.1023/A:1004155810302
Domaizon I, Savichtcheva O, Debroas D, Arnaud F, Villar C, Pignol C, Alric B, Perga ME (2013) DNA from lake sediments reveals the long-term dynamics and diversity of Synechococcus assemblages. Biogeosci Discuss 10:2515–2564. https://doi.org/10.5194/bgd-10-2515-2013
Domaizon I, Winegardner A, Capo E, Gauthier J, Gregory-Eaves I (2017) DNA-based methods in paleolimnology: new opportunities for investigating long-term dynamics of lacustrine biodiversity. J Paleolimnol 52:1–21. https://doi.org/10.1007/s10933-017-9958-y
Dräger N, Theuerkauf M, Szeroczyńska K, Wulf S, Tjallingii R, Plessen B, Kienel U, Brauer A (2017) Varve microfacies and varve preservation record of climate change and human impact for the last 6000 years at Lake Tiefer See (NE Germany). The Holocene 27:450–464. https://doi.org/10.1177/0959683616660173
Dräger N, Plessen B, Kienel U, Slowinski M, Ramisch A, Tjallingii R, Pinkerneil S, Brauer A (2019) Hypolimnetic oxygen conditions influence varve preservation and δ13C of sediment organic matter in Lake Tiefer See, NE Germany. J Paleolimnol 62:181–194. https://doi.org/10.1007/s10933-019-00084-2
Edgar RC (2010) Search and clustering hundreds of times faster than BLAST. Bioinformatics 26(19):2460–2461. https://doi.org/10.1093/bioinformatics/btq461
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron 4:1–9. https://doi.org/10.1016/j.bcp.2008.05.025
Herrera A, Cockell CS (2007) Exploring microbial diversity in volcanic environments: a review of methods in DNA extraction. J Microbiol Methods 70:1–12
Irfan S, Alatawi AMM (2019) Aquatic ecosystem and biodiversity: a review. Open J Ecol 9:1–13. https://doi.org/10.4236/oje.2019.91001
Jeppesen E, Søndergaard M, Jensen JP, Havens KE, Anneville O, Carvalho L, Coveney MF, Deneke R, Dokulil MT, Foy B, Gerdeux D, Hampton SE, Hilt S, Kangur K, Köhler J, Lammens EH, Lauridsen TL, Manca M, Miracle MR, Moss B, Noges P, Pearsson G, Phillips G, Portielje R, Romo S, Schelske CL, Straile D, Tatrai I, Willen E, Winder M (2005) Lake responses to reduced nutrient loading—an analysis of contemporary long-term data from 35 case studies. Freshw Biol 50:1747–1771. https://doi.org/10.1111/j.1365-2427.2005.01415.x
Kaiser J, Ön B, Arz H, Akçer-Ön S (2016) Sedimentary lipid biomarkers in the magnesium rich and highly alkaline lake Salda (South-western anatolia). J Limnol 75(3):581–596. https://doi.org/10.4081/jlimnol.2016.1337
Kaiser J, Wasmund N, Kahru M, Wittenborn AK, Hansen R, Häusler K, Moros M, Schulz-Bull D, Arz H (2020) Reconstructing N2-fixing cyanobacterial blooms in the Baltic Sea beyond observations using 6- And 7-methylheptadecane in sediments as specific biomarkers. Biogeosciences 17:2579–2591. https://doi.org/10.5194/bg-17-2579-2020
Kienel U, Dulski P, Ott F, Lorenz S, Brauer A (2013) Recently induced anoxia leading to the preservation of seasonal laminae in two NE-German lakes. J Paleolimnol 50:535–544. https://doi.org/10.1007/s10933-013-9745-3
Kienel U, Kirillin G, Brademann B, Plessen B, Lampe R, Brauer A (2017) Effects of spring warming and mixing duration on diatom deposition in deep Tiefer See, NE Germany. J Paleolimnol 57:37–49. https://doi.org/10.1007/s10933-016-9925-z
Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41(1):e1. https://doi.org/10.1093/nar/gks808
Komárek J, Kaštovský J, Mareš J, Johansen JR (2014) Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. Preslia 86:295–335
Legendre P, Gallagher ED (2001) Ecologically meaningful transformations for ordination of species data. Oecologia 129:271–280. https://doi.org/10.1007/s004420100716
Legendre P, Borcard D, Peres-Neto PR (2005) Analyzing beta diversity: Partitioning the spatial variation of community composition data. Ecol Monogr 75(4):435–450. https://doi.org/10.1890/05-0549
Letunic I, Bork P (2016) Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:242–245. https://doi.org/10.1093/nar/gkw290
Liu A, Zhu T, Lu X, Song L (2013) Hydrocarbon profiles and phylogenetic analyses of diversified cyanobacterial species. Appl Energy 11:383–393. https://doi.org/10.1016/j.apenergy.2013.05.008
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar BA, Lai T, Steppi S, Jobb G, Förster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, König A, Liss T, Lüßmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH (2004) ARB: A software environment for sequence data. Nucleic Acids Res 32(4):1363–1367. https://doi.org/10.1093/nar/gkh293
Martin M (2014) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. https://doi.org/10.14806/ej.17.1.200
Monchamp ME, Walser JC, Pomati F, Spaak P (2016) Sedimentary DNA reveals cyanobacterial community diversity over 200 years in two perialpine lakes. Appl Environ Microbiol 82:6472–6482. https://doi.org/10.1128/AEM.02174-16
Monchamp ME, Spaak P, Domaizon I, Dubois N, Bouffard D, Pomati F (2018) Homogenization of lake cyanobacterial communities over a century of climate change and eutrophication. Nat Ecol Evol 2(2):317–324. https://doi.org/10.1038/s41559-017-0407-0
Monchamp M-E, Spaak P, Pomati F (2019) Long term diversity and distribution of non-photosynthetic cyanobacteria in Peri-Alpine lakes. Front Microbiol 9:3344. https://doi.org/10.3389/fmicb.2018.03344
Nübel U, Garcia-Pichel F, Muyzer G (1997) PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl Environ Microbiol 63:3327–3332
Paerl HW, Huisman J (2009) Climate change: a catalyst for global expansion of harmful cyanobacterial blooms. Environ Microbiol Rep 1(1):27–37. https://doi.org/10.1111/j.1758-2229.2008.00004.x
Pfeifer F (2012) Distribution, formation and regulation of gas vesicles. Nat Rev Microbiol 10:705–715. https://doi.org/10.1038/nrmicro2834
Preußel K, Stüken A, Wiedner C, Chorus I, Fastner J (2006) First report on cylindrospermopsin producing Aphanizomenon flos-aquae (Cyanobacteria) isolated from two German lakes. Toxicon 47(2):156–62. https://doi.org/10.1016/j.toxicon.2005.10.013
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. https://doi.org/10.1093/nar/gks1219
Ramette A, Tiedje JM (2007) Multiscale responses of microbial life to spatial distance and environmental heterogeneity in a patchy ecosystem. Proc Natl Acad Sci U S A 104(8):2761–2766. https://doi.org/10.1073/pnas.0610671104
Ramos V, Morais J, Vasconcelos VM (2017) A curated database of cyanobacterial strains relevant for modern taxonomy and phylogenetic studies. Sci Data 4(1):1–8. https://doi.org/10.1038/sdata.2017.54
Rigosi A, Carey CC, Ibelings BW, Brookes JD (2014) The interaction between climate warming and eutrophication to promote cyanobacteria is dependent on trophic state and varies among taxa. Limnol Oceanogr 59:99–114. https://doi.org/10.4319/lo.2014.59.01.0099
Saary P, Forslund K, Bork P, Hildebrand F (2017) RTK: efficient rarefaction analysis of large datasets. Bioinformatics 33:2594–2595. https://doi.org/10.1093/bioinformatics/btx206
Schertzer WM, Sawchuk AM (1990) Thermal structure of the lower great lakes in a warm year: implications for the occurrence of hypolimnion anoxia. Trans Am Fish Soc 119(2):195–209. https://doi.org/10.1577/1548-8659(1990)119%3c0195
Smilauer P, Leps J (2014) Multivariate analysis of ecological data using Canoco 5, 2nd edn. Cambridge University Press, Cambridge
Smol J (2009) Pollution of lakes and rivers: a paleoenvironmental perspective. J Paleolimnol 42:301–302. https://doi.org/10.1007/s10933-009-9320-0
Soo RM, Skennerton CT, Sekiguchi Y, Imelfort M, Paech SJ, Denniss PG, Steen JA, Parks DH, Tyson GW, Hugenholtz P (2014) An expanded genomic representation of the phylum cyanobacteria. Genome Biol Evol 6(5):1031–1045. https://doi.org/10.1093/gbe/evu073
Soo RM, Hemp J, Parks DH, Fischer WW, Hugenholtz P (2017) On the origins of oxygenic photosynthesis and aerobic respiration in Cyanobacteria. Science 355(6332):1436–1440. https://doi.org/10.1126/science.aal3794
Steffen W, Crutzen PJ, McNeill JR (2007) The anthropocene: are humans now overwhelming the great forces of nature. AMBIO A J Hum Environ 36:614–621. https://doi.org/10.1579/0044-7447(2007)36[614:TAAHNO]2.0.CO;2
Sukenik A, Hadas O, Kaplan A, Quesada A (2012) Invasion of Nostocales (cyanobacteria) to subtropical and temperate freshwater lakes—physiological, regional, and global driving forces. Front Microbiol 3:86. https://doi.org/10.3389/fmicb.2012.00086
Taranu ZE, Gregory-Eaves I, Leavitt PR, Bunting L, Buchaca T, Catalan J, Domaizon I, Guilizzoni P, Lami A, McGowan S, Moohouse H, Moraboti G, Pick FR, Stevenson MA, Thompson PL, Vinebrooke RD (2015) Acceleration of cyanobacterial dominance in north temperate-subarctic lakes during the Anthropocene. Ecol Lett 18:375–384. https://doi.org/10.1111/ele.12420
Theuerkauf M, Dräger N, Kienel U, Kuparinen A, Brauer A (2015) Effects of changes in land management practices on pollen productivity of open vegetation during the last century derived from varved lake sediments. Holocene 25(5):733–744. https://doi.org/10.1177/0959683614567881
Visser PM, Ibelings BW, Bormans M, Huisman J (2016) Artificial mixing to control cyanobacterial blooms: a review. Aquat Ecol 50:423–441. https://doi.org/10.1007/s10452-015-9537-0
Wrighton KC, Castelle CJ, Wilkins MJ, Hug LA, Sharon I, Thomas BC, Handley KM, Mullin SW, Nicora CD, Singh A, Lipton MS, Long PE, Williams KH, Banfield JF (2014) Metabolic interdependencies between phylogenetically novel fermenters and respiratory organisms in an unconfined aquifer. ISME J 8(7):1452–1463. https://doi.org/10.1038/ismej.2013.249
Wulf S, Dräger N, Ott F, Serb J, Appelt O, Gudmundsdottir E, van den Bogaard C, Slowinski M, Blaszkiewicz M, Brauer A (2016) Holocene tephrostratigraphy of varved sediment records from Lakes Tiefer See (NE Germany) and Czechowskie (N Poland). Quat Sci Rev 132:1–14. https://doi.org/10.1016/j.quascirev.2015.11.007
Ye W, Tan J, Liu X, Lin S, Pan J, Li D, Yang H (2011) Temporal variability of cyanobacterial populations in the water and sediment samples of Lake Taihu as determined by DGGE and real-time PCR. Harmful Algae 10(5):472–479. https://doi.org/10.1016/j.hal.2011.03.002
Acknowledgements
We thank the German Environmental Foundation (Deutsche Bundesstiftung Umwelt DBU) for funding the research position of EN. This study is a contribution to the Virtual Institute of Integrated Climate and Landscape Evolution Analyses—ICLEA, Grant No. VH-VI-415. The monitoring equipment was funded by the Terrestrial Environmental Observatory Infrastructure Initiative of the Helmholtz Association (TERENO Observatory NE Germany). This study was also supported by the Helmholtz Gemeinschaft (HGF) by funding the Helmholtz Young Investigators Group of S.L. (VH-NG-919). JK and AB were supported by the Leibniz Association Grant SAW-2017-IOW-2. We thank the coring team of GFZ-section Climate Dynamics and Landscape Evolution who assisted in the fieldwork (B. Brademann, S. Pinkerneil) and B. Plessen for TN analyses. We thank the editors and anonymous reviewers for suggestions that improved the manuscript. We also thank L. Ganzert for help with bioinformatics and result discussions.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nwosu, E.C., Brauer, A., Kaiser, J. et al. Evaluating sedimentary DNA for tracing changes in cyanobacteria dynamics from sediments spanning the last 350 years of Lake Tiefer See, NE Germany. J Paleolimnol 66, 279–296 (2021). https://doi.org/10.1007/s10933-021-00206-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10933-021-00206-9