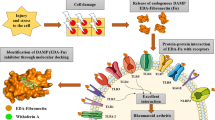
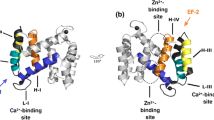
Abstract
S100A8 and S100A9 belong to the calcium-binding, damage associated molecular pattern (DAMP) proteins shown to aggravate the pathogenesis of rheumatoid arthritis (RA) through their interaction with the TLR4, RAGE and CD36 receptors. S100A8 and S100A9 proteins tend to exist in monomeric, homo and heterodimeric forms, which have been implicated in the pathogenesis of RA, via interacting with Pattern Recognition receptors (PRRs). The study aims to assess the influence of changes in the structure and biological assembly of S100A8 and S100A9 proteins as well as their interaction with significant receptors in RA through computational methods and surface plasmon resonance (SPR) analysis. Molecular docking analysis revealed that the S100A9 homodimer and S100A8/A9 heterodimer showed higher binding affinity towards the target receptors. Most S100 proteins showed good binding affinity towards TLR4 compared to other receptors. Based on the 50 ns MD simulations, TLR4, RAGE, and CD36 formed stable complexes with the monomeric and dimeric forms of S100A8 and S100A9 proteins. However, SPR analysis showed that the S100A8/A9 heterodimers formed stable complexes and exhibited high binding affinity towards the receptors. SPR data also indicated that TLR4 and its interactions with S100A8/A9 proteins may play a primary role in the pathogenesis of RA, with additional contributions from CD36 and RAGE interactions. Subsequent in vitro and in vivo investigations are warranted to corroborate the involvement of S100A8/A9 and the expression of TLR4, RAGE, and CD36 in the pathophysiology of RA.







Similar content being viewed by others
Data Availability
Data is provided within the manuscript and supplementary information files.
References
Scherer HU, Häupl T, Burmester GR (2020) The etiology of rheumatoid arthritis. J Autoimmun 102400. https://doi.org/10.1016/j.jaut.2019.102400
Kang KY, Woo J-W, Park S-H (2014) S100A8/A9 as a biomarker for synovial inflammation and joint damage in patients with rheumatoid arthritis. Korean J Intern Med 29:12–19. https://doi.org/10.3904/kjim.2014.29.1.12
Inciarte-Mundo J, Frade-Sosa B, Sanmartí R (2022) From bench to bedside: Calprotectin (S100A8/S100A9) as a biomarker in rheumatoid arthritis. Front Immunol 13:1–13
Riva M, He Z, Källberg E et al (2013) Human S100A9 protein is stabilized by inflammatory Stimuli via the formation of Proteolytically-resistant homodimers. PLoS ONE 8:e61832
Austermann J, Spiekermann C, Roth J (2018) S100 proteins in rheumatic diseases. Nat Rev Rheumatol 14:528–541. https://doi.org/10.1038/s41584-018-0058-9
Simard J-C, Cesaro A, Chapeton-Montes J et al (2013) S100A8 and S100A9 induce Cytokine expression and regulate the NLRP3 Inflammasome via ROS-Dependent activation of NF-κB1. PLoS ONE 8:e72138
Chen Y, Ouyang Y, Li Z et al (2023) S100A8 and S100A9 in Cancer. Biochim Biophys Acta - Rev Cancer 1878:188891. https://doi.org/10.1016/j.bbcan.2023.188891
Björk P, Björk A, Vogl T et al (2009) Identification of human S100A9 as a Novel Target for treatment of Autoimmune Disease via binding to Quinoline-3-Carboxamides. PLOS Biol 7:e1000097
Källberg E, Vogl T, Liberg D et al (2012) S100A9 interaction with TLR4 promotes tumor growth. PLoS ONE 7:e34207. https://doi.org/10.1371/journal.pone.0034207
Roszkowski L, Jaszczyk B, Plebańczyk M, Ciechomska M (2023) S100A8 and S100A12 proteins as biomarkers of High Disease activity in patients with rheumatoid arthritis that can be regulated by Epigenetic Drugs. Int J Mol Sci 24
Holzinger D, Tenbrock K, Roth J (2019) Alarmins of the S100-Family in Juvenile Autoimmune and Auto-Inflammatory diseases. Front Immunol 10
He Z, Riva M, Björk P et al (2016) CD14 is a Co-receptor for TLR4 in the S100A9-Induced pro-inflammatory response in Monocytes. PLoS ONE 11:e0156377
C (2022) Role of the S100 protein family in rheumatoid arthritis. Arthritis Res Ther 24:35. https://doi.org/10.1186/s13075-022-02727-8
Polakowska M, Steczkiewicz K, Szczepanowski RH, Wysłouch-Cieszyńska A (2023) Toward an understanding of the conformational plasticity of S100A8 and S100A9 Ca2+-binding proteins. J Biol Chem 299. https://doi.org/10.1016/j.jbc.2023.102952
Bresnick AR (2018) S100 proteins as therapeutic targets. Biophys Rev 10:1617–1629. https://doi.org/10.1007/s12551-018-0471-y
Li D, Wu M (2021) Pattern recognition receptors in health and diseases. Signal Transduct Target Ther 6:291. https://doi.org/10.1038/s41392-021-00687-0
Heo Y-J, Oh H-J, Jung YO et al (2011) The expression of the receptor for advanced glycation end-products (RAGE) in RA-FLS is induced by IL-17 via Act-1. Arthritis Res Ther 13:R113. https://doi.org/10.1186/ar3398
Bongarzone S, Savickas V, Luzi F, Gee AD (2017) Targeting the receptor for Advanced Glycation endproducts (RAGE): a Medicinal Chemistry Perspective. J Med Chem 60:7213–7232. https://doi.org/10.1021/acs.jmedchem.7b00058
Ma L, Sun P, Zhang J-C et al (2017) Proinflammatory effects of S100A8/A9 via TLR4 and RAGE signaling pathways in BV-2 microglial cells. Int J Mol Med 40:31–38. https://doi.org/10.3892/ijmm.2017.2987
Volz HC, Laohachewin D, Seidel C et al (2012) S100A8/A9 aggravates post-ischemic heart failure through activation of RAGE-dependent NF-κB signaling. Basic Res Cardiol 107:250. https://doi.org/10.1007/s00395-012-0250-z
Chen Y, Zhang J, Cui W, Silverstein RL (2022) CD36, a signaling receptor and fatty acid transporter that regulates immune cell metabolism and fate. J Exp Med 219:e20211314. https://doi.org/10.1084/jem.20211314
Wang S, Song R, Wang Z et al (2018) S100A8/A9 in inflammation. Front Immunol 9
Kim H-A, Han JH, Kim W-J et al (2016) TLR4 endogenous ligand S100A8/A9 levels in adult-onset still’s disease and their association with Disease Activity and Clinical manifestations. Int J Mol Sci 17
Kuipers MT, Vogl T, Aslami H et al (2013) High levels of S100A8/A9 proteins aggravate Ventilator-Induced Lung Injury via TLR4 Signaling. PLoS ONE 8:e68694
Tan X, Zheng X, Huang Z et al (2017) Involvement of S100A8/A9-TLR4-NLRP3 inflammasome pathway in contrast-Induced Acute kidney Injury. Cell Physiol Biochem 43:209–222. https://doi.org/10.1159/000480340
Narumi K, Miyakawa R, Ueda R et al (2015) Proinflammatory proteins S100A8/S100A9 activate NK cells via Interaction with RAGE. J Immunol 194:5539–5548. https://doi.org/10.4049/jimmunol.1402301
Hermani A, De Servi B, Medunjanin S et al (2006) S100A8 and S100A9 activate MAP kinase and NF-κB signaling pathways and trigger translocation of RAGE in human prostate cancer cells. Exp Cell Res 312:184–197. https://doi.org/10.1016/j.yexcr.2005.10.013
Ghavami S, Rashedi I, Dattilo BM et al (2008) S100A8/A9 at low concentration promotes tumor cell growth via RAGE ligation and MAP kinase-dependent pathway. J Leukoc Biol 83:1484–1492. https://doi.org/10.1189/jlb.0607397
Kerkhoff C, Sorg C, Tandon NN, Nacken W (2001) Interaction of S100A8/S100A9-arachidonic acid complexes with the scavenger receptor CD36 may facilitate fatty acid uptake by endothelial cells. Biochemistry 40:241–248. https://doi.org/10.1021/bi001791k
Joshi A, Schmidt LE, Burnap SA et al (2022) Neutrophil-derived protein S100A8/A9 alters the platelet proteome in Acute myocardial infarction and is Associated with changes in platelet reactivity. Arterioscler Thromb Vasc Biol 42:49–62. https://doi.org/10.1161/ATVBAHA.121.317113
Wang Y, Fang C, Gao H et al (2014) Platelet-derived S100 family member myeloid-related protein-14 regulates thrombosis. J Clin Invest 124:2160–2171. https://doi.org/10.1172/JCI70966
Szklarczyk D, Gable AL, Nastou KC et al (2021) The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 49:D605–D612. https://doi.org/10.1093/nar/gkaa1074
Shannon P, Markiel A, Ozier O et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/gr.1239303
Schrödinger LLC (2015) The {PyMOL} Molecular Graphics System, Version ~ 1.8
Kozakov D, Hall DR, Xia B et al (2017) The ClusPro web server for protein–protein docking. Nat Protoc 12:255–278. https://doi.org/10.1038/nprot.2016.169
van Zundert GCP, Rodrigues JPGLM, Trellet M et al (2016) The HADDOCK2.2 web server: user-friendly integrative modeling of Biomolecular complexes. J Mol Biol 428:720–725. https://doi.org/10.1016/j.jmb.2015.09.014
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25(13):1605–1612. https://doi.org/10.1002/jcc.20084
Xue LC, Rodrigues JP, Kastritis PL et al (2016) PRODIGY: a web server for predicting the binding affinity of protein–protein complexes. Bioinformatics 32:3676–3678. https://doi.org/10.1093/bioinformatics/btw514
Abraham MJ, Murtola T, Schulz R et al (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1:19–25. https://doi.org/10.1016/j.softx.2015.06.001
Bjelkmar P, Larsson P, Cuendet MA et al (2010) Implementation of the CHARMM Force Field in GROMACS: analysis of Protein Stability effects from correction maps, Virtual Interaction Sites, and Water models. J Chem Theory Comput 6:459–466. https://doi.org/10.1021/ct900549r
Turner PJ (2005) XMGRACE, Version 5.1. 19. Cent Coast Land-Margin Res Oregon Grad Inst Sci Technol Beaverton, OR 2
Mehmood A, Nawab S, Jia G et al (2023) Supervised screening of Tecovirimat-like compounds as potential inhibitors for the monkeypox virus E8L protein. J Biomol Struct Dyn 1–14. https://doi.org/10.1080/07391102.2023.2245042
Mehmood A, Nawab S, Jin Y et al (2023) Ranking breast Cancer drugs and biomarkers Identification using machine learning and Pharmacogenomics. ACS Pharmacol Transl Sci 6:399–409. https://doi.org/10.1021/acsptsci.2c00212
Mehmood A, Nawab S, Jin Y et al (2023) Mutational impacts on the N and C terminal domains of the MUC5B protein: a transcriptomics and Structural Biology Study. ACS Omega 8:3726–3735. https://doi.org/10.1021/acsomega.2c04871
Mehmood A, Kaushik AC, Wang Q et al (2021) Bringing structural implications and deep learning-based drug identification for KRAS mutants. J Chem Inf Model 61:571–586. https://doi.org/10.1021/acs.jcim.0c00488
Chen B, Miller AL, Rebelatto M et al (2015) S100A9 Induced inflammatory responses are mediated by distinct damage Associated Molecular patterns (DAMP) receptors in Vitro and in vivo. PLoS ONE 10:e0115828
Edgeworth J, Gorman M, Bennett R et al (1991) Identification of p8,14 as a highly abundant heterodimeric calcium binding protein complex of myeloid cells. J Biol Chem 266:7706–7713. https://doi.org/10.1016/S0021-9258(20)89506-4
Prantner D, Nallar S, Vogel SN (2020) The role of RAGE in host pathology and crosstalk between RAGE and TLR4 in innate immune signal transduction pathways. FASEB J 34:15659–15674. https://doi.org/10.1096/fj.202002136R
Möller A, Jauch-Speer S-L, Gandhi S et al (2023) The roles of toll-like receptor 4, CD33, CD68, CD69, or CD147/EMMPRIN for monocyte activation by the DAMP S100A8/S100A9. Front. Immunol. 14
Sunahori K, Yamamura M, Yamana J et al (2006) The S100A8/A9 heterodimer amplifies proinflammatory cytokine production by macrophages via activation of nuclear factor kappa B and p38 mitogen-activated protein kinase in rheumatoid arthritis. Arthritis Res Ther 8:R69. https://doi.org/10.1186/ar1939
Colicchia M, Schrottmaier WC, Perrella G et al (2022) S100A8/A9 drives the formation of procoagulant platelets through GPIbα. Blood 140:2626–2643. https://doi.org/10.1182/blood.2021014966
Cao D, Luo J, Chen D et al (2016) CD36 regulates lipopolysaccharide-induced signaling pathways and mediates the internalization of Escherichia coli in cooperation with TLR4 in goat mammary gland epithelial cells. Sci Rep 6:23132. https://doi.org/10.1038/srep23132
Shmuel-Galia L, Klug Y, Porat Z et al (2017) Intramembrane attenuation of the TLR4-TLR6 dimer impairs receptor assembly and reduces microglia-mediated neurodegeneration. J Biol Chem 292:13415–13427. https://doi.org/10.1074/jbc.M117.784983
Chávez-Sánchez L, Garza-Reyes MG, Espinosa-Luna JE et al (2014) The role of TLR2, TLR4 and CD36 in macrophage activation and foam cell formation in response to oxLDL in humans. Hum Immunol 75:322–329. https://doi.org/10.1016/j.humimm.2014.01.012
Kim HM, Park BS, Kim J-I et al (2007) Crystal structure of the TLR4-MD-2 complex with bound Endotoxin Antagonist Eritoran. Cell 130:906–917. https://doi.org/10.1016/j.cell.2007.08.002
Edilova MI, Akram A, Abdul-Sater AA (2021) Innate immunity drives pathogenesis of rheumatoid arthritis. Biomed J 44:172–182. https://doi.org/10.1016/j.bj.2020.06.010
Qin W, Rong X, Yu C et al (2022) Knockout of SLAMF8 attenuates collagen-induced rheumatoid arthritis in mice through inhibiting TLR4/NF-κB signaling pathway. Int Immunopharmacol 107:108644. https://doi.org/10.1016/j.intimp.2022.108644
Sun X, Zhang T, Liu S et al (2023) The prepared and characterized polysaccharide polymer in Saposhnikovia divaricata(Trucz.) Schischk effectively controls the course of rheumatoid arthritis via TLR4/TRAF6–NF-κB/IκB-α signaling pathway. Biomed Pharmacother 160:114416. https://doi.org/10.1016/j.biopha.2023.114416
Li L, Pan Z, Ning D, Fu Y (2022) Rosmanol and Carnosol Synergistically Alleviate Rheumatoid Arthritis through Inhibiting TLR4/NF-κB/MAPK Pathway. Molecules 27.
Zheng J, Wang J, Liu H et al (2022) Alarmins S100A8/A9 promote intervertebral disc degeneration and inflammation-related pain in a rat model through toll-like receptor-4 and activation of the NF-κB signaling pathway. Osteoarthr Cartil 30:998–1011. https://doi.org/10.1016/j.joca.2022.03.011
Grevers LC, De Vries TJ, Vogl T et al (2011) S100A8 enhances osteoclastic bone resorption in vitro through activation of toll-like receptor 4: implications for bone destruction in murine antigen-induced arthritis. Arthritis Rheum 63. https://doi.org/10.1002/art.30290
Zhong A, Xu W, Zhao J et al (2016) S100A8 and S100A9 are Induced by decreased hydration in the Epidermis and promote fibroblast activation and fibrosis in the Dermis. Am J Pathol 186:109–122. https://doi.org/10.1016/j.ajpath.2015.09.005
Wang L, Luo H, Chen X et al (2014) Functional characterization of S100A8 and S100A9 in altering monolayer permeability of human umbilical endothelial cells. PLoS ONE 9:e90472
Ostrand-Rosenberg S, Huecksteadt T, Sanders K (2023) The receptor for Advanced Glycation endproducts (RAGE) and its ligands S100A8/A9 and high mobility Group Box protein 1 (HMGB1) are key regulators of myeloid-derived suppressor cells. Cancers (Basel). 15
Gheibi N, Ghorbani M, Shariatifar H, Farasat A (2019) In silico assessment of human calprotectin subunits (S100A8/A9) in presence of sodium and calcium ions using Molecular Dynamics simulation approach. PLoS ONE 14:e0224095. https://doi.org/10.1371/journal.pone.0224095
Acknowledgements
The authors would like to acknowledge SPR Facility, National Centre for Cell Science (NCCS), Pune, for the facility and assistance provided throughout the study.
Funding
This work was supported by the Indian Council of Medical Research (ICMR), New Delhi (Sanction order No: F. No. 58/12/2020/PHA/BMS Dtd: 04/03/2022).
Author information
Authors and Affiliations
Contributions
Methodology, formal analysis, Data collection, interpretation, writing - S.P.; conceptualization, critical revision of the manuscript – S.S.P.; Conceptualization, critical revision of the manuscript, supervision of the study, funding acquisition – S.P.E. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Paramasivam, S., Perumal, S.S. & Ekambaram, S.P. Computational Deciphering of the Role of S100A8 and S100A9 Proteins and Their Changes in the Structure Assembly Influences Their Interaction with TLR4, RAGE, and CD36. Protein J (2024). https://doi.org/10.1007/s10930-024-10186-0
Accepted:
Published:
DOI: https://doi.org/10.1007/s10930-024-10186-0