Abstract
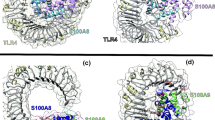
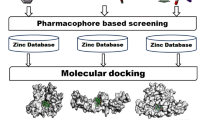
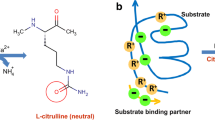
Rheumatoid arthritis (RA) is one of the most severe inflammatory diseases that cause swelling, stiffness and pain in the joints, which pose a significant threat worldwide. Damage-associated molecular patterns (DAMPs) are danger molecules of endogenous origin, released during cell injury or cell death, interacts with various Pattern recognition receptors (PRRs) and activates various inflammatory diseases. One of the DAMP molecules, so-called EDA-fibronectin (Fn) is also responsible for causing RA. EDA-Fn triggers RA through its interaction with TLR4. Apart from TLR4, it is divulged that certain other PRR’s are also responsible for RA, but the identity and mechanism of those PRRs remain unknown until now. Hence, for the first time, we tried to reveal those PRR’s interaction with EDA-Fn in RA through computational methods. Protein–protein interaction (PPI) was checked using ClusPro between EDA-Fn and certain Pattern recognition receptors (PRRs) to explore the binding affinities of the potential PRRs. Protein–protein docking unveiled that TLR5, TLR2 and RAGE has good interaction with EDA-Fn than the well-reported TLR4. Macromolecular simulation was performed for TLR5, TLR2 and RAGE complexes along with the control group TLR4 for 50 ns to further investigate the stability, leading to the identification of TLR2, TLR5 and RAGE as the stable complexes. Hence, TLR2, TLR5 and RAGE on interaction with EDA-Fn may lead to the progression of RA that may need additional validations through in vitro and in vivo animal models. Molecular docking was used to analyse the binding force of the top 33 active anti-arthritic compounds with the target protein EDA-Fn. Molecular docking study showed that withaferin A has a good binding activity with EDA-fibronectin target. Hence, it is emphasized that guggulsterone and berberine could modulate the EDA-Fn-mediated TLR5/TLR2/RAGE pathways, thereby it could inhibit the deteriorating effects of RA which needs further in vitro and in vivo experimental validations.
Graphical Abstract












Similar content being viewed by others
Data Availability
Data will be available on request.
References
Guo, Q., Wang, Y., Xu, D., Nossent, J., Pavlos, N. J., & Xu, J. (2018). Rheumatoid arthritis: Pathological mechanisms and modern pharmacologic therapies. Bone Research, 6(1), 15. https://doi.org/10.1038/s41413-018-0016-9
Matcham, F., Scott, I. C., Rayner, L., Hotopf, M., Kingsley, G. H., Norton, S., & Steer, S. (2014). The impact of rheumatoid arthritis on quality-of-life assessed using the SF-36: A systematic review and meta-analysis. Seminars in Arthritis and Rheumatism, 44(2), 123–130. https://doi.org/10.1016/j.semarthrit.2014.05.001
Edwards, C. J., & Cooper, C. (2006). Early environmental factors and rheumatoid arthritis. Clinical and Experimental Immunology, 143(1), 1–5. https://doi.org/10.1111/j.1365-2249.2005.02940.x
Bullock, J., Rizvi, S. A. A., Saleh, A. M., Ahmed, S. S., Do, D. P., Ansari, R. A., & Ahmed, J. (2019). Rheumatoid arthritis: A brief overview of the treatment. Medical Principles and Practice, 27(6), 501–507. https://doi.org/10.1159/000493390
Tanaka, Y., Millson, D., Iwata, S., & Nakayamada, S. (2021). Safety and efficacy of fostamatinib in rheumatoid arthritis patients with an inadequate response to methotrexate in phase II OSKIRA-ASIA-1 and OSKIRA-ASIA-1X study. Rheumatology (United Kingdom), 60(6), 2884–2895. https://doi.org/10.1093/rheumatology/keaa732
Rudan, I., Sidhu, S., Papana, A., Meng, S. J., Xin-Wei, Y., Wang, W., Chan, K. Y., & Global Health Epidemiology Reference Group (GHERG) (2015). Prevalence of rheumatoid arthritis in low- and middle-income countries: A systematic review and analysis. Journal of Global Health, 5(1). https://doi.org/10.7189/jogh.05.010409
Gerosa, M., De Angelis, V., Riboldi, P., & Meroni, P. L. (2008). Rheumatoid arthritis: A female challenge. Women’s Health, 4(2), 195–201. https://doi.org/10.2217/17455057.4.2.195
Alamanos, Y., Voulgari, P. V., & Drosos, A. A. (2006). Incidence and prevalence of rheumatoid arthritis, based on the 1987 American College of Rheumatology criteria: A systematic review. Seminars in Arthritis and Rheumatism, 36(3), 182–188. https://doi.org/10.1016/J.SEMARTHRIT.2006.08.006
Roh, J. S., & Sohn, D. H. (2018). Origin and List of DAMPS. Immune Network, 18(4), 1–14.
Navrátilová, A., Bečvář, V., Baloun, J., Damgaard, D., Nielsen, C. H., Veigl, D., & Andrés Cerezo, L. (2021). S100A11 (calgizzarin) is released via NETosis in rheumatoid arthritis (RA) and stimulates IL-6 and TNF secretion by neutrophils. Scientific Reports, 11(1), 1–11. https://doi.org/10.1038/s41598-021-85561-3
Huang, Q.-Q., Sobkoviak, R., Jockheck-Clark, A. R., Shi, B., Mandelin, A. M., Tak, P. P., & Pope, R. M. (2009). Heat shock protein 96 Is elevated in rheumatoid arthritis and activates macrophages primarily via TLR2 signaling. The Journal of Immunology, 182(8), 4965–4973. https://doi.org/10.4049/jimmunol.0801563
Shiozawa, K., Hino, K., & Shinozawa, S. (2001). Alternatively spliced EDA-containing fibronectin in synovial fluid as a predictor of rheumatoid joint destruction. Rheumatology, 40(7), 739–742. https://doi.org/10.1093/rheumatology/40.7.739
Sun, V. Z., Melim, T. L., Mitra, S., Erickson, J. E., Bryant, S. H., Farnham, A., Goodearl, A. D. (2022). Fibronectin extra domain A as a drug delivery targeting epitope for rheumatoid arthritis. Advances in Rheumatology, 62(1):17. https://doi.org/10.1186/s42358-022-00247-2
Nawaz, H., Ali, A., Rehman, T., & Aslam, A. (2021). Chronological effects of non-steroidal anti-inflammatory drug therapy on oxidative stress and antioxidant status in patients with rheumatoid arthritis. Clinical Rheumatology, 40(5), 1767–1778. https://doi.org/10.1007/s10067-020-05438-0
Wang, W., Zhou, H., & Liu, L. (2018). Side effects of methotrexate therapy for rheumatoid arthritis: A systematic review. European Journal of Medicinal Chemistry, 158, 502–516. https://doi.org/10.1016/j.ejmech.2018.09.027
Gondokaryono, S. P., Ushio, H., Niyonsaba, F., Hara, M., Takenaka, H., Jayawardana, S. T. M., & Ogawa, H. (2007). The extra domain A of fibronectin stimulates murine mast cells via Toll-like receptor 4. Journal of Leukocyte Biology, 82(3), 657–665. https://doi.org/10.1189/jlb.1206730
Chang, X., Yamada, R., Suzuki, A., Kochi, Y., Sawada, T., & Yamamoto, K. (2005). Citrullination of fibronectin in rheumatoid arthritis synovial tissue. Rheumatology, 44(11), 1374–1382. https://doi.org/10.1093/rheumatology/kei023
Shelef, M. A., Bennin, D. A., Mosher, D. F., & Huttenlocher, A. (2012). Citrullination of fibronectin modulates synovial fibroblast behavior. Arthritis Research and Therapy, 14, R240. https://doi.org/10.1186/ar4083
Huang, Q.-Q., & Pope, R. M. (2009). The role of toll-like receptors in rheumatoid arthritis. Current Rheumatology Reports, 11(5), 357–364.
Mullen, L. M., Chamberlain, G., & Sacre, S. (2015). Pattern recognition receptors as potential therapeutic targets in inflammatory rheumatic disease. Arthritis Research and Therapy, 17(1), 1–10. https://doi.org/10.1186/s13075-015-0645-y
Gonzalez, M. W., & Kann, M. G. (2012). Chapter 4: Protein interactions and disease. PLoS Computational Biology, 8(12): e1002819. https://doi.org/10.1371/journal.pcbi.1002819
Ryan, D. P., & Matthews, J. M. (2005). Protein-protein interactions in human disease. Current Opinion in Structural Biology, 15(4), 441–446. https://doi.org/10.1016/j.sbi.2005.06.001
Rajendran, V., Purohit, R., & Sethumadhavan, R. (2012). In silico investigation of molecular mechanism of laminopathy caused by a point mutation (R482W) in lamin A/C protein. Amino Acids, 43(2), 603–615. https://doi.org/10.1007/s00726-011-1108-7
Rajendran, V., & Sethumadhavan, R. (2014). Drug resistance mechanism of PncA in Mycobacterium tuberculosis. Journal of Biomolecular Structure and Dynamics, 32(2), 209–221. https://doi.org/10.1080/07391102.2012.759885
Rajendran, V. (2016). Structural analysis of oncogenic mutation of isocitrate dehydrogenase 1. Molecular BioSystems, 12(7), 2276–2287. https://doi.org/10.1039/c6mb00182c
Kumar, S., Bhardwaj, V. K., Singh, R., Das, P., & Purohit, R. (2022). Identification of acridinedione scaffolds as potential inhibitor of DENV-2 C protein: An in silico strategy to combat dengue. Journal of Cellular Biochemistry, 123(5), 935–946. https://doi.org/10.1002/jcb.30237
Rajendran, V., Gopalakrishnan, C., & Purohit, R. (2016). Impact of point mutation P29S in RAC1 on tumorigenesis. Tumor Biology, 37(11), 15293–15304. https://doi.org/10.1007/s13277-016-5329-y
Rajendran, V., Gopalakrishnan, C., & Sethumadhavan, R. (2018). Pathological role of a point mutation (T315I) in BCR-ABL1 protein—A computational insight. Journal of Cellular Biochemistry, 119(1), 918–925. https://doi.org/10.1002/jcb.26257
Singh, R., Bhardwaj, V. K., Das, P., & Purohit, R. (2022). Identification of 11β-HSD1 inhibitors through enhanced sampling methods. Chemical Communications, 58(32), 5005–5008. https://doi.org/10.1039/D1CC06894F
Khan, A. U., Khan, A., Khan, A., Shal, B., Aziz, A., Ahmed, M. N., & Khan, S. (2021). Inhibition of NF-κB signaling and HSP70/HSP90 proteins by newly synthesized hydrazide derivatives in arthritis model. Naunyn-Schmiedeberg’s Archives of Pharmacology, 394(7), 1497–1519. https://doi.org/10.1007/s00210-021-02075-5
Zou, X., Yang, X. J., Gan, Y. M., Liu, D. L., Chen, C., Duan, W., & Du, J. R. (2021). Neuroprotective effect of phthalide derivative CD21 against ischemic brain injury:Involvement of MSR1 mediated DAMP peroxiredoxin1 clearance and TLR4 signaling inhibition. Journal of Neuroimmune Pharmacology, 16(2), 306–317. https://doi.org/10.1007/s11481-020-09911-0
Choudhary, M., Kumar, V., Malhotra, H., & Singh, S. (2015). Medicinal plants with potential anti-arthritic activity. Journal of Intercultural Ethnopharmacology, 4(2), 147. https://doi.org/10.5455/jice.20150313021918
Niimi, T., Osawa, M., Yamaji, N., Yasunaga, K., Sakashita, H., Mase, T., & Fujita, S. (2001). Letter to the Editor: NMR structure of human fibronectin EDA. Journal of Biomolecular NMR, 21(3), 281–284. https://doi.org/10.1023/A:1012947209393
Jin, M. S., Kim, S. E., Heo, J. Y., Lee, M. E., Kim, H. M., Paik, S.-G., & Lee, J.-O. (2007). Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell, 130(6), 1071–1082. https://doi.org/10.1016/j.cell.2007.09.008
Bell, J. K., Botos, I., Hall, P. R., Askins, J., Shiloach, J., Segal, D. M., & Davies, D. R. (2005). The molecular structure of the Toll-like receptor 3 ligand-binding domain. Proceedings of the National Academy of Sciences, 102(31), 10976–10980. https://doi.org/10.1073/pnas.0505077102
Park, B. S., Song, D. H., Kim, H. M., Choi, B.-S., Lee, H., & Lee, J.-O. (2009). The structural basis of lipopolysaccharide recognition by the TLR4–MD-2 complex. Nature, 458(7242), 1191–1195. https://doi.org/10.1038/nature07830
Zhou, K., Kanai, R., Lee, P., Wang, H.-W., & Modis, Y. (2012). Toll-like receptor 5 forms asymmetric dimers in the absence of flagellin. Journal of Structural Biology, 177(2), 402–409. https://doi.org/10.1016/j.jsb.2011.12.002
Jang, T., & Park, H. H. (2014). Crystal structure of TIR domain of TLR6 reveals novel dimeric interface of TIR–TIR interaction for Toll-like receptor signaling pathway. Journal of Molecular Biology, 426(19), 3305–3313. https://doi.org/10.1016/j.jmb.2014.07.024
Ishida, H., Asami, J., Zhang, Z., Nishizawa, T., Shigematsu, H., Ohto, U., & Shimizu, T. (2021). Cryo-EM structures of Toll-like receptors in complex with UNC93B1. Nature Structural & Molecular Biology, 28(2), 173–180. https://doi.org/10.1038/s41594-020-00542-w
Xu, Y., Li, W., Ke, H., & Feng, W. (2018). Structural conservation of the autoinhibitory domain in SUN proteins. Biochemical and Biophysical Research Communications, 496(4), 1337–1343. https://doi.org/10.1016/j.bbrc.2018.02.015
Yatime, L., & Andersen, G. R. (2013). Structural insights into the oligomerization mode of the human receptor for advanced glycation end-products. The FEBS Journal, 280(24), 6556–6568. https://doi.org/10.1111/febs.12556
Hsieh, F.-L., Turner, L., Bolla, J. R., Robinson, C. V., Lavstsen, T., & Higgins, M. K. (2016). The structural basis for CD36 binding by the malaria parasite. Nature Communications, 7(1), 12837. https://doi.org/10.1038/ncomms12837
Dekker, C., Mattes, H., Wright, M., Boettcher, A., Hinniger, A., Hughes, N., & Farady, C. J. (2021). Crystal structure of NLRP3 NACHT domain with an inhibitor defines mechanism of inflammasome inhibition. Journal of Molecular Biology, 433(24), 167309. https://doi.org/10.1016/j.jmb.2021.167309
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., & Ferrin, T. E. (2004). UCSF Chimera - A visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25(13), 1605–1612. https://doi.org/10.1002/jcc.20084
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., & Schwede, T. (2018). SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Research, 46(W1), W296–W303. https://doi.org/10.1093/nar/gky427
Hollingsworth, S. A., & Karplus, P. A. (2010). A fresh look at the Ramachandran plot and the occurrence of standard structures in proteins. Biomolecular Concepts, 1(3–4), 271–283. https://doi.org/10.1515/bmc.2010.022
Desta, I. T., Porter, K. A., Xia, B., Kozakov, D., & Vajda, S. (2020). Performance and its limits in rigid body protein-protein docking. Structure, 28(9), 1071-1081.e3. https://doi.org/10.1016/j.str.2020.06.006
Kozakov, D., Hall, D. R., Xia, B., Porter, K. A., Padhorny, D., Yueh, C., & Vajda, S. (2017). The ClusPro web server for protein-protein docking. Nature Protocols, 12(2), 255–278. https://doi.org/10.1038/nprot.2016.169
Pierce, B. G., Wiehe, K., Hwang, H., Kim, B. H., Vreven, T., & Weng, Z. (2014). ZDOCK server: Interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics, 30(12), 1771–1773. https://doi.org/10.1093/bioinformatics/btu097
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., & Ferrin, T. E. (2004). UCSF Chimera—A visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25(13), 1605–1612. https://doi.org/10.1002/jcc.20084
Laskowski, R. A., & Swindells, M. B. (2011). LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. Journal of Chemical Information and Modeling, 51(10), 2778–2786. https://doi.org/10.1021/ci200227u
Abraham, M. J., Murtola, T., Schulz, R., Páll, S., Smith, J. C., Hess, B., & Lindahl, E. (2015). GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX, 1, 19–25. https://doi.org/10.1016/j.softx.2015.06.001
Turner, P. J. (2005). XMGRACE, version 5.1. 19. center for coastal and land-margin research, Oregon Graduate Institute of Science and Technology, Beaverton, OR; 2005
Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S., & Bolton, E. E. (2021). PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Research, 49(D1), D1388–D1395. https://doi.org/10.1093/nar/gkaa971
O’Boyle, N. M., Banck, M., James, C. A., Morley, C., Vandermeersch, T., & Hutchison, G. R. (2011). Open Babel. Journal of Cheminformatics, 3(33), 1–14.
Kutzner, C., Páll, S., Fechner, M., Esztermann, A., de Groot, B. L., & Grubmüller, H. (2019). More bang for your buck: Improved use of GPU nodes for GROMACS 2018. Journal of Computational Chemistry, 40(27), 2418–2431. https://doi.org/10.1002/jcc.26011
van Aalten, D. M. F., Bywater, R., Findlay, J. B. C., Hendlich, M., Hooft, R. W. W., & Vriend, G. (1996). PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. Journal of Computer-Aided Molecular Design, 10(3), 255–262. https://doi.org/10.1007/BF00355047
Pol-Fachin, L., Fernandes, C. L., & Verli, H. (2009). GROMOS96 43a1 performance on the characterization of glycoprotein conformational ensembles through molecular dynamics simulations. Carbohydrate Research, 344(4), 491–500. https://doi.org/10.1016/j.carres.2008.12.025
Premnath, S., Indrajith, S., Senthamil Selvan, P., & Sanmuga Priya, E. (2022). The role of fibronectin and its isoforms in the pathogenesis and progression of rheumatoid arthritis: A review. Biointerface Research in Applied Chemistry, 13(4), 341. https://doi.org/10.33263/BRIAC134.341
Roberts, A. L., Mavlyutov, T. A., Perlmutter, T. E., Curry, S. M., Harris, S. L., Chauhan, A. K., & McDowell, C. M. (2020). Fibronectin extra domain A (FN-EDA) elevates intraocular pressure through Toll-like receptor 4 signaling. Scientific Reports, 10(1), 9815. https://doi.org/10.1038/s41598-020-66756-6
Sargsyan, K., Grauffel, C., & Lim, C. (2017). How molecular size impacts RMSD applications in molecular dynamics simulations. Journal of Chemical Theory and Computation, 13(4), 1518–1524. https://doi.org/10.1021/acs.jctc.7b00028
Shtaiwi, A., Adnan, R., Khairuddean, M., & Al-Qattan, M. (2018). Molecular dynamics simulation of human estrogen receptor free and bound to morpholine ether benzophenone inhibitor. Theoretical Chemistry Accounts, 137(7), 101. https://doi.org/10.1007/s00214-018-2277-1
Seibl, R., Birchler, T., Loeliger, S., Hossle, J. P., Gay, R. E., Saurenmann, T., & Lauener, R. P. (2003). Expression and regulation of Toll-like receptor 2 in rheumatoid arthritis synovium. The American Journal of Pathology, 162(4), 1221–1227. https://doi.org/10.1016/S0002-9440(10)63918-1
Oliviera Nascimento, L., Massari, P., & Wetzler, L. (2012). The role of TLR2 in infection and immunity. Frontiers in Immunology, 3:79. https://doi.org/10.3389/fimmu.2012.00079
Chamberlain, N. D., Vila, O. M., Volin, M. V., Volkov, S., Pope, R. M., Swedler, W., & Shahrara, S. (2012). TLR5, a novel and unidentified inflammatory mediator in rheumatoid arthritis that correlates with disease activity score and joint TNF-α levels. Journal of Immunology, 189(1), 475–483. https://doi.org/10.4049/jimmunol.1102977
Prantner, D., Nallar, S., & Vogel, S. N. (2020). The role of RAGE in host pathology and crosstalk between RAGE and TLR4 in innate immune signal transduction pathways. The FASEB Journal, 34(12), 15659–15674. https://doi.org/10.1096/fj.202002136R
Author information
Authors and Affiliations
Contributions
Premnath Sakthivel: data curation, formal analysis, methodology, software, visualisation, writing—original draft, writing—review and editing. Indrajith Sakthivel: Data curation, formal analysis, methodology, software, visualisation, writing—review and editing. Sivasakthi Paramasivam: data curation, methodology, software, visualisation, writing—review and editing. Senthamil Selvan Perumal: data curation, methodology, software, supervision, writing—review and editing. Sanmuga Priya Ekambaram: conceptualization, investigation, project administration, supervision, validation, writing—review and editing.
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
All authors read and approved the manuscript for publication.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sakthivel, P., Sakthivel, I., Paramasivam, S. et al. Underpinning Endogeneous Damp EDA-Fibronectin in the Activation of Molecular Targets of Rheumatoid Arthritis and Identifcation of its Effective Inhibitors by Computational Methods. Appl Biochem Biotechnol 195, 7037–7059 (2023). https://doi.org/10.1007/s12010-023-04451-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-023-04451-8