Abstract
Monkeypox, a viral zoonotic disease resembling smallpox, has emerged as a significant national epidemic primarily in Africa. Nevertheless, the recent global dissemination of this pathogen has engendered apprehension regarding its capacity to metamorphose into a sweeping pandemic. To effectively combat this menace, a multi-epitope vaccine has been meticulously engineered with the specific aim of targeting the cell envelope protein of Monkeypox virus (MPXV), thereby stimulating a potent immunological response while mitigating untoward effects. This new vaccine uses T-cell and B-cell epitopes from a highly antigenic, non-allergenic, non-toxic, conserved, and non-homologous A30L protein to provide protection against the virus. In order to ascertain the vaccine design with the utmost efficacy, protein–protein docking methodologies were employed to anticipate the intricate interactions with Toll-like receptors (TLR) 2, 3, 4, 6, and 8. This meticulous approach led the researchers to discern an optimal vaccine architecture, bolstered by affirmative prognostications derived from both molecular dynamics (MD) simulations and immune simulations. The current research findings indicate that the peptides ATHAAFEYSK, FFIVVATAAV, and MNSLSIFFV exhibited antigenic properties and were determined to be non-allergenic and non-toxic. Through the utilization of codon optimization and in-silico cloning techniques, our investigation revealed that the prospective vaccine exhibited a remarkable expression level within Escherichia coli. Moreover, upon conducting immune simulations, we observed the induction of a robust immune response characterized by elevated levels of both B-cell and T-cell mediated immunity. Moreover, as the initial prediction with in-silico techniques has yielded promising results these epitope-based vaccines can be recommended to in vitro and in silico studies to validate their immunogenic properties.













Similar content being viewed by others
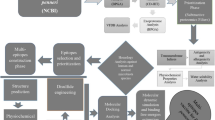
Availability of Data And Material
Not Applicable.
Code Availability
Not Applicable.
Abbreviations
- ANN:
-
Artificial Neural Network
- CAI:
-
Codon Adaptation Index
- HCID:
-
High-Consequence Infectious Diseases
- MHC I:
-
Major Histocompatibility Complex I
- MHC II:
-
Major Histocompatibility Complex II
- MPXV:
-
Monkey Pox Virus
- PSSM:
-
Position Specific Scoring Matrix
- SASA:
-
Solvent Accessible Surface Area
- TLR:
-
Toll Like Receptor
References
Shafi A, Wani AA, Ul Islam J, Peerzada K (2022) Monkey pox: What we need to know. Acta Sci Microbiol. https://doi.org/10.31080/asmi.2022.05.1095
von Magnus P, Andersen EK, Petersen KB, Birch-Andersen A (2009) A pox-like disease in Cynomolgus monkeys. Acta Pathol Microbiol Scand 46(2):156–176. https://doi.org/10.1111/j.1699-0463.1959.tb00328.x
Ladnyj ID, Ziegler P, Kima E (1972) A human infection caused by monkeypox virus in Basankusu Territory, Democratic Republic of the Congo. Bull World Health Organ 46(5):593–597
Likos AM, Sammons SA, Olson VA, Frace AM, Li Y, Olsen-Rasmussen M et al (2005) A tale of two clades: monkeypox viruses. J Gen Virol 86(Pt 10):2661–2672. https://doi.org/10.1099/vir.0.81215-0
Zumla A, Valdoleiros SR, Haider N, Asogun D, Ntoumi F, Petersen E et al (2022) Monkeypox outbreaks outside endemic regions: scientific and social priorities. Lancet Infect Dis 22(7):929–931. https://doi.org/10.1016/S1473-3099(22)00354-1
Titanji BK, Tegomoh B, Nematollahi S, Konomos M, Kulkarni PA (2022) Monkeypox: A contemporary review for healthcare professionals. Open Forum Infect Dis 9(7):ofac310. https://doi.org/10.1093/ofid/ofac310
Jain N, Lansiaux E, Simanis R (2022) The new face of monkeypox virus: an emerging global emergency. New Microbes New Infect. 47(100989):100989. https://doi.org/10.1016/j.nmni.2022.100989
Kumar R, Nagar S, Haider S, Sood U, Ponnusamy K, Dhingra GG, et al. Monkey pox virus (MPXV): Phylogenomics, host-pathogen interactome, and mutational cascade. bioRxiv. 2022. doi:https://doi.org/10.1101/2022.07.25.501367
Daskalakis D, Mcclung RP, Mena L, Mermin J (2022) Centers for Disease Control and Prevention’s Monkeypox Response Team. Ann Intern Med 175(8):1177–1178
Islam MR, Hossain MJ, Roy A, Hasan AHMN, Rahman MA, Shahriar M et al (2022) Repositioning potentials of smallpox vaccines and antiviral agents in monkeypox outbreak: A rapid review on comparative benefits and risks. Health Sci Rep. 5(5):e798. https://doi.org/10.1002/hsr2.798
Shchelkunov SN, Totmenin AV, Safronov PF, Mikheev MV, Gutorov VV, Ryazankina OI et al (2002) Analysis of the monkeypox virus genome. Virology 297(2):172–194. https://doi.org/10.1006/viro.2002.1446
Kozlova EEG, Cerf L, Schneider FS, Viart BT, NGuyen C, Steiner BT et al (2018) Computational B-cell epitope identification and production of neutralizing murine antibodies against Atroxlysin-I. Sci Rep 8(1):14904. https://doi.org/10.1038/s41598-018-33298-x
Chauhan V, Rungta T, Goyal K, Singh MP (2019) Designing a multi-epitope-based vaccine to combat Kaposi Sarcoma utilizing immunoinformatics approach. Sci Rep 9(1):2517. https://doi.org/10.1038/s41598-019-39299-8
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD et al (2005) Protein identification and analysis tools on the ExPASy server. In: Handbook TPP (ed) Totowa. Humana Press, NJ, pp 571–607
Doytchinova IA, Flower DR (2007) Identifying candidate subunit vaccines using an alignment-independent method based on principal amino acid properties. Vaccine 25(5):856–866. https://doi.org/10.1016/j.vaccine.2006.09.032
Doytchinova IA, Flower DR (2007) VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics 8(1):4. https://doi.org/10.1186/1471-2105-8-4
Gupta S, Kapoor P, Chaudhary K, Gautam A, Kumar R, Open Source Drug Discovery Consortium et al (2013) In silico approach for predicting toxicity of peptides and proteins. PLoS ONE 8(9):e73957. https://doi.org/10.1371/journal.pone.0073957
Tarek MM, Shafei AE, Ali MA, Mansour MM (2018) Computational prediction of vaccine potential epitopes and 3-dimensional structure of XAGE-1b for non-small cell lung cancer immunotherapy. Biomed J 41(2):118–128. https://doi.org/10.1016/j.bj.2018.04.002
Dimitrov I, Bangov I, Flower DR, Doytchinova I (2014) AllerTOP vol 2–a server for in silico prediction of allergens. J Mol Model 20(6):2278. https://doi.org/10.1007/s00894-014-2278-5
Wang P, Sidney J, Dow C, Mothé B, Sette A, Peters B (2008) A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol 4(4):e1000048. https://doi.org/10.1371/journal.pcbi.1000048
Jespersen MC, Peters B, Nielsen M, Marcatili P (2017) BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res 45(W1):W24–W29. https://doi.org/10.1093/nar/gkx346
Nielsen M, Lundegaard C, Worning P, Lauemøller SL, Lamberth K, Buus S et al (2003) Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci 12(5):1007–1017. https://doi.org/10.1110/ps.0239403
Lundegaard C, Lamberth K, Harndahl M, Buus S, Lund O, Nielsen M (2008) NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res 36(suppl_2):509–512. https://doi.org/10.1093/nar/gkn202
Kolaskar AS, Tongaonkar PC (1990) A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett 276(1–2):172–174. https://doi.org/10.1016/0014-5793(90)80535-q
Singh A, Thakur M, Sharma LK, Chandra K (2020) Designing a multi-epitope peptide based vaccine against SARS-CoV-2. Sci Rep 10(1):16219. https://doi.org/10.1038/s41598-020-73371-y
Jensen KK, Andreatta M, Marcatili P, Buus S, Greenbaum JA, Yan Z et al (2018) Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology 154(3):394–406. https://doi.org/10.1111/imm.12889
Bui H-H, Sidney J, Dinh K, Southwood S, Newman MJ, Sette A (2006) Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinformatics 7(1):153. https://doi.org/10.1186/1471-2105-7-153
Kar T, Narsaria U, Basak S, Deb D, Castiglione F, Mueller DM et al (2020) A candidate multi-epitope vaccine against SARS-CoV-2. Sci Rep 10(1):10895. https://doi.org/10.1038/s41598-020-67749-1
Naz A, Shahid F, Butt TT, Awan FM, Ali A, Malik A (2020) Designing multi-Epitope vaccines to combat emerging Coronavirus disease 2019 (COVID-19) by employing immuno-informatics approach. Front Immunol 11:1663. https://doi.org/10.3389/fimmu.2020.01663
Samad A, Ahammad F, Nain Z, Alam R, Imon RR, Hasan M et al (2022) Designing a multi-epitope vaccine against SARS-CoV-2: an immunoinformatics approach. J Biomol Struct Dyn 40(1):14–30. https://doi.org/10.1080/07391102.2020.1792347
Kouza M, Faraggi E, Kolinski A, Kloczkowski A (2017) The GOR method of protein secondary structure prediction and its application as a protein aggregation prediction tool. Methods Mol Biol 1484:7–24. https://doi.org/10.1007/978-1-4939-6406-2_2
Roy A, Kucukural A, Zhang Y (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5(4):725–738. https://doi.org/10.1038/nprot.2010.5
Yang J, Zhang Y (2015) Protein structure and function prediction using I-TASSER. Curr Protoc Bioinformatics 52(1):5.8.1-5.8.15. https://doi.org/10.1002/0471250953.bi0508s52
Ferrè F, Clote P (2005) DiANNA: a web server for disulfide connectivity prediction. Nucleic Acids Res 33(Web Server issue):W230–W232. https://doi.org/10.1093/nar/gki412
Yan Y, Tao H, He J, Huang S-Y (2020) The HDOCK server for integrated protein-protein docking. Nat Protoc 15(5):1829–1852. https://doi.org/10.1038/s41596-020-0312-x
Yan Y, Zhang D, Zhou P, Li B, Huang S-Y (2017) HDOCK: a web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res 45(W1):W365–W373. https://doi.org/10.1093/nar/gkx407
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B et al (2015) GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2:19–25. https://doi.org/10.1016/j.softx.2015.06.001
Abraham MJ, Gready JE (2011) Optimization of parameters for molecular dynamics simulation using smooth particle-mesh Ewald in GROMACS 4.5. J Comput Chem 32(9):2031–2040. https://doi.org/10.1002/jcc.21773
Mauro VP (2018) Codon optimization in the production of recombinant biotherapeutics: Potential risks and considerations. BioDrugs 32(1):69–81. https://doi.org/10.1007/s40259-018-0261-x
Castiglione F, Bernaschi M. C-immsim: playing with the immune response. In: Proceedings of the sixteenth international symposium on mathematical theory of networks and systems (MTNS2004). Belgium; 2004.
Rapin N, Lund O, Bernaschi M, Castiglione F (2010) Computational immunology meets bioinformatics: the use of prediction tools for molecular binding in the simulation of the immune system. PLoS ONE 5(4):e9862. https://doi.org/10.1371/journal.pone.0009862
Shilling PJ, Mirzadeh K, Cumming AJ, Widesheim M, Köck Z, Daley DO (2020) Improved designs for pET expression plasmids increase protein production yield in Escherichia coli. Commun Biol 3(1):214. https://doi.org/10.1038/s42003-020-0939-8
Ortiz-Saavedra B, León-Figueroa DA, Montes-Madariaga ES, Ricardo-Martínez A, Alva N, Cabanillas-Ramirez C et al (2022) Antiviral treatment against Monkeypox: A scoping review. Trop Med Infect Dis 7(11):369. https://doi.org/10.3390/tropicalmed7110369
Lim HX, Lim J, Jazayeri SD, Poppema S, Poh CL (2021) Development of multi-epitope peptide-based vaccines against SARS-CoV-2. Biomed J 44(1):18–30. https://doi.org/10.1016/j.bj.2020.09.005
Romeli S, Hassan SS, Yap WB (2020) Multi-epitope peptide-based and vaccinia-based universal influenza vaccine candidates subjected to clinical trials. The Malaysian journal of medical sciences MJMS. 27(2):10–20
Zhang L (2018) Multi-epitope vaccines: a promising strategy against tumors and viral infections. Cell Mol Immunol 15(2):182–184. https://doi.org/10.1038/cmi.2017.92
Yadav P, Devasurmutt Y, Tatu U (2022) Phylogenomic and structural analysis of the Monkeypox virus shows evolution towards increased stability. Viruses 15(1):127. https://doi.org/10.3390/v15010127
Arai R, Ueda H, Kitayama A, Kamiya N, Nagamune T (2001) Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng 14(8):529–532. https://doi.org/10.1093/protein/14.8.529
Matsumoto M, Oshiumi H, Seya T (2011) Antiviral responses induced by the TLR3 pathway: antiviral function of TLR3. Rev Med Virol 21(2):67–77. https://doi.org/10.1002/rmv.680
Zhang S-Y, Herman M, Ciancanelli MJ, Pérez de Diego R, Sancho-Shimizu V, Abel L et al (2013) TLR3 immunity to infection in mice and humans. Curr Opin Immunol 25(1):19–33. https://doi.org/10.1016/j.coi.2012.11.001
Chen R (2012) Bacterial expression systems for recombinant protein production: E. coli and beyond. Biotechnol Adv 30(5):1102–1107. https://doi.org/10.1016/j.biotechadv.2011.09.013
Rosano GL, Ceccarelli EA (2014) Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol 5:172. https://doi.org/10.3389/fmicb.2014.00172
Shantier SW, Mustafa MI, Abdelmoneim AH, Fadl HA, Elbager SG, Makhawi AM (2022) Novel multi epitope-based vaccine against monkeypox virus: vaccinomic approach. Sci Rep 12(1):15983. https://doi.org/10.1038/s41598-022-20397-z
Abdi SAH, Ali A, Sayed SF, Abutahir AA, Alam P (2022) Multi-Epitope-based vaccine candidate for Monkeypox: An in silico approach. Vaccines (Basel). 10(9):1564. https://doi.org/10.3390/vaccines10091564
Zaib S, Rana N, Areeba HN, Alrbyawi H, Dera AA et al (2023) Designing multi-epitope monkeypox virus-specific vaccine using immunoinformatics approach. J Infect Public Health 16(1):107–116. https://doi.org/10.1016/j.jiph.2022.11.033
Bhattacharya K, Shamkh IM, Khan MS, Lotfy MM, Nzeyimana JB, Abutayeh RF et al (2022) Multi-Epitope vaccine design against Monkeypox virus via reverse vaccinology method exploiting immunoinformatic and bioinformatic approaches. Vaccines (Basel) 10(12):2010. https://doi.org/10.3390/vaccines10122010
Khan S, Irfan M, Hameed AR, Ullah A, Abideen SA, Ahmad S et al (2022) Vaccinomics to design a multi-epitope-based vaccine against monkeypox virus using surface-associated proteins. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2022.2158942
Ullah A, Shahid FA, Haq MU, Tahir Ul Qamar M, Irfan M, Shaker B, et al. An integrative reverse vaccinology, immunoinformatic, docking and simulation approaches towards designing of multi-epitopes based vaccine against monkeypox virus. J Biomol Struct Dyn. 2022;1–14. doi:https://doi.org/10.1080/07391102.2022.2125441
Rcheulishvili N, Mao J, Papukashvili D, Feng S, Liu C, Yang X et al (2023) Development of a multi-Epitope universal mRNA vaccine candidate for Monkeypox, smallpox, and Vaccinia viruses: Design and in silico analyses. Viruses 15(5):1120. https://doi.org/10.3390/v15051120
Jahantigh HR, Shahbazi B, Gouklai H, Van der Weken H, Gharibi Z, Rezaei Z et al (2023) Design peptide and multi-epitope protein vaccine candidates against monkeypox virus using reverse vaccinology approach: an in-silico study. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2023.2201850
Lahimchi MR, Madanchi H, Ahmadi K, Shahbazi B, Yousefi B (2023) In silico designing a novel TLR4-mediating multiepitope vaccine against monkeypox via advanced immunoinformatics and bioinformatics approaches. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2023.2203253
Novitsky V, Flores-Villanueva PO, Chigwedere P, Gaolekwe S, Bussman H, Sebetso G et al (2001) Identification of most frequent HLA class I antigen specificities in Botswana: relevance for HIV vaccine design. Hum Immunol 62(2):146–156. https://doi.org/10.1016/s0198-8859(00)00236-6
Acknowledgements
The authors would like to acknowledge the Bioinformatics lab facility, BioNome (https://bionome.in/), Bangalore, 560043, India, for their assistance in the work. The help from Dr. Sameer Sharma and his team, Bioinformatics lab, was greatly helpful in performing MD simulation analysis via Gromacs v2019.4.
Funding
Not Applicable.
Author information
Authors and Affiliations
Contributions
All the authors have equally contributed to the manuscript.
Corresponding author
Ethics declarations
Conflicts of Interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ramprasadh, S.V., Rajakumar, S., Srinivasan, S. et al. Computer-Aided Multi-Epitope Based Vaccine Design Against Monkeypox Virus Surface Protein A30L: An Immunoinformatics Approach. Protein J 42, 645–663 (2023). https://doi.org/10.1007/s10930-023-10150-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-023-10150-4



