Abstract
COVID-19 is a disease that have affected the entire world, and it continues to spread with new variants. A patient’s innate immune system plays a critical role in the mild and severe transition of COVID-19. Antimicrobial peptides (AMPs), which are important components of the innate immune system, are potential molecules to fight pathogenic bacteria, fungi, and viruses. Human β-defensin 2 (hBD-2), a 41-amino-acid antimicrobial peptide, is one of the defensins inducibly expressed in the skin, lungs, and trachea in humans. In this study, it was aimed to investigate the interaction of hBD-2 produced recombinantly in Pichia pastoris with the human angiotensin-converting enzyme 2 (ACE-2) under in vitro conditions. First, hBD-2 was cloned in P. pastoris X-33 via the pPICZαA vector, a yeast expression platform, and its expression was confirmed by SDS-PAGE, western blotting, and qRT-PCR. Then, the interaction between recombinant hBD-2 and ACE-2 proteins was revealed by a pull-down assay. In light of these preliminary experiments, we suggest that the recombinantly produced hBD-2 may be protective against SARS-CoV-2 and be used as a supplement in treatment. However, current findings need to be supported by cell culture studies, toxicity analyses, and in vivo experiments.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Antimicrobial peptides (AMPs) are short peptide molecules containing 10–100 amino acids, also called host defense peptides (HDP), produced by most living things, and generally rich in cationic amino acids like lysine and arginine. AMPs are considered to be among new-generation and multifunctional antibiotics that can be a solution to antimicrobial resistance and viral infections [1, 2].
Defensins, one of the essential groups of AMPs, can be a good alternative for protecting the host against microbial infections, while also limiting the increase in antimicrobial resistance, which has become a global health problem [3]. ß-defensins, which are effective against Gram-negative and Gram-positive bacteria, cause multiple stresses either by acting on bacterial membranes or by affecting DNA synthesis and repair, protein synthesis, and cell signal transduction pathways. One of the most essential characteristics of ß-defensins is that they develop low bacterial resistance due to their antibacterial properties and show minimal toxic or allergic effects on humans [3, 4]. In addition to antimicrobial resistance, many studies have shown that AMPs could be used as an effective agent against SARS-CoV-2 at the beginning of the COVID-19 pandemic.
COVID-19 is an infectious viral disease first detected in China in 2019 [5, 6]. SARS-CoV-2 is enveloped by single-stranded ribonucleic acid as its nuclear material. It expresses the spike (S) protein, which interacts with the angiotensin-converting enzyme-2 (ACE-2) receptors of respiratory and digestive epithelial cells. The initial stages of SARS-CoV-2 infection concern the specific binding of the S protein of SARS-CoV-2 to ACE-2 [7, 8]. This binding induces a conformational change in S proteins. After this, proteolytic degradation occurs via proteases on the cell surface. This way, the virion can enter the cell [9]. Due to this pathway that is seen during the entry of the infectious agent, the inhibition of both the S protein and ACE-2 protein is a promising approach in the potential treatment process [10]. ACE-2 gene expression was reported in the lungs, liver, brain, testes, kidneys, and heart. ACE-2 has many different functions as a cell surface receptor in these organs [11].
Human β-defensin 2 (hBD-2) is a low-molecular-weight AMP discovered in the human skin in 1997 [12]. hBD-2 is transcribed from the Defb4 gene, has 41 amino acids and is inducibly expressed in the skin lesions of psoriasis patients and the lung epithelium of cystic fibrosis patients [12, 13]. hBD-2 is one of the AMP molecules currently being reevaluated. Recent studies have reported that hBD-2 blocks the receptor-binding domain (RBD) of the S protein in in silico and cell cultures [14, 15]. However, the mechanism of this inhibition process is still unclear.
Recently, along with the COVID-19 pandemic, the overuse of antibiotics and the spread of hospital infections have led to an increased interest in AMP molecules [16]. On the other hand, for AMPs to be used in clinical settings, they must be produced in large amounts. AMP extraction from natural sources is laborious and substantially affected by environmental conditions. Another method is to produce AMPs by chemical synthesis. However, chemical synthesis from long peptide sequences is not preferred due to the difficulty of the method and its increased costs [17, 18]. Because of these disadvantages, recombinant approaches are currently preferred for the production of AMPs. Additionally, it is biotechnologically significant to ensure that recombinant proteins can be produced on a large scale, and their therapeutic properties are maintained [17, 18].
In this study, we aimed to produce hBD-2 recombinantly in Pichia pastoris X-33, demonstrate its activity, and examine whether it binds to the ACE-2 receptor under in vitro conditions. This way, we wanted to provide preliminary data on whether recombinantly produced hBD-2 can bind with ACE-2 and have a protective effect.
2 Materials and methods
2.1 Reagents, Plasmids and Pathogenic Bacterial Strains
pPICZαA and the P. pastoris X-33 strain were used for cloning studies. Zeocin was purchased from InvivoGen (Toulouse, France). Restriction endonucleases were obtained from New England Biolabs (Ipswich, United States). A plasmid extraction, gel purification kit and SDS-PAGE marker was obtained from EcoTech Biotechnology (Erzurum, Turkey). For qRT-PCR, HOT FIREPol® EvaGreen® qPCR Supermix (Solis BioDyne, Estonia) was used. The MagneHis™ Protein Purification System cat number V8500 was purchased from Promega Co. Ltd. (Madison, USA). ACE-2 protein (cat no: AVI10544) was utilized, and a pull-down assay kit (Pierce™ His Protein Interaction Pull-Down Kit) was purchased from Thermo Scientific (Rockford, USA). The purity of other chemical reagents was of analytical grade. All primers that were used in the study are given in Table 1.
2.2 Construction Design, Cloning, and Transformation
The coding region of hBD-2 was excised from the FaDu cell line (HTB-43™) by the induction method with some modifications [19] (Fig.S1). In this process, the FaDu cell line was incubated with 1% L-glutamine, 1% penicillin/streptomycin, and 10% (/v) fetal calf serum in RPMI 1640 media. The flask was incubated at 37 °C with 5% CO2 in the incubator. At the same time, Pseudomonas aeruginosa (ATCC 10,145) was inoculated in LB broth medium at 37 °C and 150 rpm. After the adhesion of the FaDu cell line, 1 ml culture filtrate of filter-sterilized P. aeruginosa was added to the cultures and incubated for 24 h. Thus, hBD-2 production was facilitated. The FaDu cell line not supplemented with P. aeruginosa culture filtrate was used as the control group. The levels of hBD-2 expression were also determined by qRT-PCR.
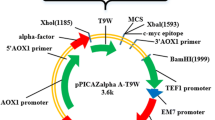
In the primer design process, the coding sequence for hBD-2 was incorporated with the EcoRI and XbaI restriction sites. The PCR product was ligated into pGEM T-easy vector and transformed into E. coli TOP10 cells. The pGEM-hBD2 vector isolated from the transformants with the help of the plasmid isolation kit was sequenced using the SP6 and T7 universal primers, and the presence of hBD-2 was confirmed. After confirmation, the pGEM-hBD2 and pPICZαA vectors were digested with the EcoRI and XbaI restriction enzymes, and the purified DNA fragments from the gel were ligated together. The resulting plasmid (pPICZaA-hBD-2) was transformed into E. coli TOP10 cells cultivated at 37℃ in low-salt LB agar medium containing 25 µg/ml of zeocin. The selected transformants were confirmed by colony PCR. Lastly, the pPICZαA-hBD2 vector was linearized with SacI to transfer electrocompetent P. pastoris X-33 cells. The electroporation (Biorad, Gene Pulser Xcell Electroporation Systems) conditions were 2000 V, 25 µF, and 200 Ω. DNA was isolated from the P. pastoris transformants using the phenol/chloroform extraction methods previously described by Sambrook and Green (2018) [20]. The linearized pPICZαA vector was used as the negative control. Transformants were selected from genomic DNA by PCR using the 5’AOX and 3’AOX primers. The construction diagram is presented in Fig.S2.
2.3 Expression and Purification of Recombinant Protein
The positive transformant was grown in 25 ml buffered glycerol complex medium (BMGY; 2% peptone, 1.4% yeast nitrogen base, 1% yeast extract, 1% glycerol, 0.00004% biotin, and 100 mM potassium phosphate buffer pH 6.0) in a 250-ml triple baffled flask at 30℃ and 150 rpm for 16 h. Then, the culture was centrifuged and washed with 5 ml of buffered methanol complex medium (BMMY; BMGY medium containing 0.5% v/v methanol instead of glycerol) to remove the remaining glycerol. To induce hBD-2 expression, the culture was started in 100 ml BMMY medium with an optical density (OD600 nm) of 1.0. Methanol induction was performed by forming three different groups containing 1%, 2%, and 3% methanol. The methanol induction process was carried out every 24 h. One ml of the sample was collected every day in all groups.
2.4 Antimicrobial Activity Assay
The P. aeruginosa (ATCC10145, PAO1), Klebsiella pneumoniae (clinical isolate oxa-48+), Acinetobacter baumannii (ATCC BAA-1605), Staphylococcus aureus (ATCC 43,300, ATCC 25,923), E. coli (ATCC 25,922), and Enterococcus faecalis (ATCC 29,212) strains were used to determine the antimicrobial activity of hBD-2.
Antimicrobial activity was evaluated using two different methods. In the first method, the collected culture filtrates were evaluated with the agar well diffusion method [21]. In this process, the pathogenic bacterial strains were seeded on Mueller Hinton agar by the spread plate method. The wells were drilled with a 6 mm cork borer, and culture filtrates were added to the wells. The specimens were incubated at 37℃ for 24 h. At the end of the incubation time, zone formations were examined. In the second method, the examination was made using a micro liquid-bacterial colony notation assay [22, 23]. Here, the overnight bacteria culture was adjusted to be an OD 600 of 1.0, and 2 µl of this suspension was taken to a 96-well plate. Then, 100-µl recombinant culture filtrates were added to a 96-well plate. BMMY medium was used as the negative control. This plate was incubated at 37 °C for 4 h. After incubation, 30 µl of the specimen was taken and spread on LB agar medium, followed by incubation at 37 °C for 24 h [23]. A colony count was performed after incubation.
2.5 RNA Isolation and cDNA Synthesis
RNA isolation was performed in the group showing antibacterial activity using the standard TRIzol-based method. Then, RNA concentration and purity were measured with the help of an Epoch Microplate Spectrophotometer (BioTek Instruments, Winooski, VT, USA), and the samples were stored at -80℃ until being used in experiments. To determine the levels of hBD-2 expression, 1 µg of RNA sample was converted to cDNA following the manufacturer’s kit protocol (Invitrogen, USA). The cDNAs formed as a result of the reaction were stored at -20 °C.
2.6 Real-Time PCR (qRT-PCR) Analysis
The resulting cDNAs were diluted at a ratio of 1:3. The qRT-PCR reaction was facilitated using the HOT FIREPol® EvaGreen® qPCR Supermix (Solis BioDyne, Estonia). The GAPDH gene was used as a reference control gene, and the experiments were performed in triplicates. The results were analyzed using the 2−(ΔΔCt) method. The significance of the results was evaluated with one-way analysis of variance (ANOVA) in the GraphPad (Prism 8.2) program.
2.7 Purification of hBD-2 and SDS-PAGE
hBD-2, which was secreted into the culture medium, was purified using the MagneHis™ Protein Purification System as described in the manufacturer’s instructions. Its concentration was determined by the Bradford assay method. To confirm the purity of hBD-2, SDS-PAGE was performed according to Brunelle and Green (2014) with some modifications [24]. The purified protein was loaded onto 15% polyacrylamide gels at a constant voltage (70 V) and identified by Coomassie blue staining. Images were taken using BioRad Gel Doc XR Imaging Systems with image lab software.
2.8 Western Blot Analysis
Equal amounts of protein samples separated in 15% SDS-PAGE gel were transferred to a ClearBand Polyvinylidene Fluoride (PVDF) membrane in a western blot system. Afterwards, they were washed with 1X TBST. The membrane was blocked using a blocking buffer for 1 h at room temperature. The primary antibody was diluted with the blocking buffer according to the dilution factor recommended in the manufacturer’s protocol. His tag primary antibody diluted enough to remove the blocking buffer and cover the membrane was added. The membrane was incubated with the primary antibody for 1 day at 4℃. The membrane was then washed three times for 15 min with 1xTBST to remove the primary antibody. The HRP-conjugated secondary antibody was diluted with the blocking buffer. The membrane was incubated with the secondary antibody for 1 h at room temperature, and it was then washed 3 times for 15 min with 1xTBST. Equal amounts of solution 1 and 2 substrate mixtures were used for chemiluminescent irradiation. The membrane was placed on a flat surface, and 1 ml of the substrate mixture for the membrane was distributed over it. After 3 min, the membrane was placed inside the Gel Doc XR Imaging Systems (BioRad), and the images were taken.
2.9 In silico Analysis
First, the amino acid sequences of human ACE-2 and hBD-2 proteins were accessed from the UniProt database (https://www.uniprot.org/). The InterEvDock2 software (https://mobyle.rpbs.univ-paris-diderot.fr/cgi-bin/portal.py#data::overview) was used to predict the binding patterns of the two sequences. The interaction was facilitated using the FRODOCK-Interactive protein-protein docking (https://frodock.iqfr.csic.es/) software. The structure was analyzed in the PyMOL program.
2.10 Pull Down Assay
The interaction assay was performed using the Pierce™ His Protein Interaction Pull-Down Kit according to the manufacturer’s recommendations. In the process, the purified hBD-2 protein was bound to the pull-down column system. To prevent unwanted binding on the column, washes were performed with a wash buffer (TBS: 25 mM Tris-Cl; 0.15 M NaCl; pH 7.2), and the wash solutions (bait flow) were collected. Then, 50 µg of ACE-2 protein was added, washed on this column for 2 h with 800 µl of an equilibration buffer (Lysis:TBS: Imidazole; 4 ml lysis buffer: 4 ml TBS:20 µl 4 M Imidazole), and incubated for 2 h at 4℃ by stirring. At the end of the process, this column was washed to remove unwanted binding. Each wash was carried out for 10 min by stirring. When the washes were completed, the samples were collected with 250 µl of elution buffer (500 mM imidazole). SDS-PAGE was performed with the samples, and the interaction was observed.
3 Results
3.1 Construction of pPICZαA-hBD-2 Vector and Selection of Recombinant Clones
The hBD-2 peptide encoded by the Defb4 gene is secreted from epithelial cells to assist host defense. Its production is induced by pathogens, endotoxins, and cytokines [25]. We set up a system called induction to easily extract the mRNA of hBD-2 from FaDu cells. Therefore, the purpose of induction is to amplify hBD-2 more easily. This way, the coding sequence of hBD-2 was amplified from the FaDu cell line, a hypopharyngeal carcinoma cell line. As shown in Fig.S3a, a 3335-fold increase in the level of hBD-2 gene expression was recorded in the induction process with P. aeruginosa (ATCC10145) compared to the control.
After this, the resulting PCR product (Fig.S3b) was first ligated into the pGEM-T easy vector and subcloned into E. coli cells. After sequence validation, the target gene region was ligated into the pPICZαA vector. Eventually, the recombinant pPICZαA-hBD-2 was confirmed by Sanger sequencing.
After the confirmation of the construct, pPICZαA-hBD-2 was linearized with SacI and transformed into the electrocompetent P. pastoris X-33 by electroporation. The recombinant clones were screened in zeocin-containing YPDS medium. A total of eleven clones were selected. Their genomic DNA was amplified by PCR using the 5’AOX and 3’AOX primers. The pPICZαA vector was used as the negative control.
3.2 Expression of Recombinant hBD-2 and Optimization of Its Production
In this study, we performed the production process by methanol induction. It is reported that the methanol ratio used in the induction process varies according to the protein [26, 27]. Therefore, the rate of methanol added in the induction process can have a positive or negative effect on production. Since the addition of methanol in excess will have a toxic effect on recombinant P. pastoris, different rates have been tested to determine the optimum methanol ratio for production. Therefore, in this study, methanol concentrations of 1–3% were used in the methanol induction process. Firstly, the selected recombinant colonies grown in the BMMY medium were induced every 24 h in different groups containing 1%, 2%, and 3% methanol. The methanol concentration at which recombinant hBD-2 was determined was 2%. When we used the other methanol ratios, we could not observe the active production of hBD-2.
qRT-PCR and western blotting experiments were performed to confirm production on the mRNA and protein levels, respectively. Here, in the examination performed with qRT-PCR, it was determined that the mRNA level was the highest at the 72nd hour as shown in Fig.S4a. hBD-2 was detected by western blotting using His-tag antibodies, and it was successfully imaged up to about 7 kDa at the 96th hour, as shown in Fig.S4b. Additionally, we obtained approximately 200 µg of protein from 100 ml of culture.
3.3 Antibacterial Activity of hBD-2
In the agar diffusion test, the culture filtrate taken at the 96th hour caused zone formation (Fig.S5). The best results were obtained against P. aeruginosa, A. baumannii, and K. pneumoniae strains with 2% methanol induction. Using this assay, we could roughly remark that the recombinant hBD-2 in the culture filtrate was active and had antimicrobial activity. Additionally, both the culture supernatant and the purified recombinant protein showed antibacterial activity against E. coli, P. aeruginosa, S. aureus, K. pneumoniae, A. baumannii, and E. faecalis, as shown in Fig.S6.
The same conditions were used in the micro liquid-bacterial colony notation assay, which evaluated the activity of the recombinant hBD-2 protein to inhibit the growth of S. aureus and P. aeruginosa as shown in Fig.S7. In this test, the examination of surviving colonies of bacteria after hBD-2 exposure was performed. Consequently, it was observed that the culture filtrates at the 96th hour of induction with 2% methanol against pathogenic bacteria significantly reduced the number of colonies. This way, recombinant hBD-2 was shown to inhibit both Gram-positive and Gram-negative bacteria.
3.4 Interaction hBD-2 and ACE-2 Proteins
In the in silico analysis, the human ACE-2 (Q9BYF1) and hBD-2 (O15263) amino acid sequences were obtained from the UniProt database. Then, the interaction was analyzed for binding patterns with the InterEvDock2 and Frodock2 software. According to the analysis results, it was bioinformatically estimated that tyrosine, lysine, glutamine, and isoleucine, which are respectively the 24th, 25th, 26th, and 27th amino acids of the hBD-2 protein and cysteine, which is the 15th amino acid of the hBD-2 protein, interacted with asparagine and tyrosine as respectively the 51st and 52nd amino acids of the ACE-2 receptor, methionine and asparagine as respectively the 62nd and 63rd amino acids of this receptor, and serine as the 47th amino acid of this receptor (Fig. 1a). The results of the analyses with the InterEvDock2 software also supported the results presented in Fig. 1b.
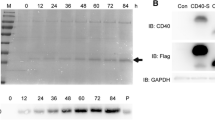
a.In silico analysis of hBD-2 and ACE-2 protein interactions. The interface between the ACE-2 and hBD-2. b. 3-D interaction InterEvDock 2.0 image c. SDS-PAGE analysis of pull down assay using recombinant hBD-2 produced from P. pastoris X-33 as the bait and ACE-2 protein as the targets. Protein bands were visualized by staining with Coomassie Blue. The image was taken with the Gel Doc XR Imaging Systems
In the pull-down assay, hBD-2 that was recombinantly produced form P. pastoris as a bait and human ACE-2 protein as a target were used. We found that the hBD-2 recombinantly produced by P. pastoris was pulled down by ACE-2 (Fig. 1c.)
4 Discussion
AMPs are a part of the immune system in multicellular organisms while helping unicellular organisms compete for nutrients against other organisms that share their biological niche. They are also the first line of defense against infectious agents. Since AMPs are very important in antibiotic resistance and antiviral treatments, their recombinant production is required.
The high chemical synthesis cost of AMPs dramatically limits their application in human diseases. Chemical synthesis costs more than $100 for 1 mg of product [28]. The recombinant production of AMPs has been attempted in different hosts over the recent years. In this regard, the synthesis of long-chain AMPs can be realized at a lower cost. E. coli, Bacillus subtilis, Saccharomyces cerevisiae, and P. pastoris have been broadly used to generate recombinant AMPs. Among these, P. pastoris is significant in that it can provide convenient production for post-translational modification such as folding, disulfide bond formation, and glycosylation. Additionally, high-density cell growth, the high efficiency of secreted proteins, and lesser amounts of immunogenic glycans are among the advantages of the P. pastoris expression system [29]. In particular, P. pastoris is preferred in the recombinant production of proteins of human origin, and P. pastoris has been successfully used for the production of more than 500 biopharmaceutical products [30]. The production of many AMP molecules has been carried out in the yeast P. pastoris as the host [31,32,33].
hBD-2 is drawing attention as an alternative therapeutic molecule for both antibiotic resistance and viral infections [34, 35]. Currently, the production pathway of hBD-2 involves chemical synthesis and heterologous expression. However, the chemical synthesis of hBD-2 is very costly, and its recombinant production involves processes that need to be optimized [36]. In this study, we produced hBD-2 using the yeast P. pastoris X-33 as the host. We confirmed its activity against pathogenic bacterial strains (Fig.S5-7).
We used the inducible promoter in the pPICZαA vector. Although the use of constitutive promoters provides advantages in the production of recombinant proteins, it is not preferred in the production of antimicrobial proteins [37]. Antimicrobial proteins can be toxic to the host cell due to their functional properties. Therefore, inducible promoters are more efficient in AMP production.
In previous studies, it has been reported that hBD-2 can be produced by prokaryotic and eukaryotic expression systems. Fang et al. (2005) [38] reported the expression of hBD-2 in E. coli as a fusion protein. Peng et al. (2004) [39] utilized a different design for the production of hBD-2 in E. coli [27]. In their study, the hBD-2 gene was cloned into the pET-32a(+) vector by performing codon optimization. They also optimized the production temperature, IPTG concentration, and medium content. Xu et al. (2006) [40] produced hBD-2 in multiple copies in E. coli. Another approach to the recombinant production of hBD-2 was introduced in 2007 by Aerts et al. [32, 41]. In their study, Arabidopsis thaliana was used as the host organism. Møller et al. (2017) [42] carried out the production of hBD-2 with the MET17 promoter in S. cerevisiae [30]. Corrales-García et al. (2020) [36] facilitated the expression of hBD-2 in E. coli using a factorial experimental design. They investigated the effect of cell density, temperature, length of induction, and inducer concentration. Consequently, the best expression condition in E. coli was determined.
In a recently published study examining the anti-cancer properties of hBD-2, it was stated that hBD-2 was produced in the P. pastoris GS115 yeast strain [43]. However, in the aforementioned study, information about antibacterial activity, production efficiency, and protein amount was not provided. Although some researchers have produced recombinant hBD-2 production using S. cerevisiae, P. pastoris, and E. coli as hosts, as far as we know, we report here for the first time the recombinant production and activity of hBD-2 using a wild-type P. pastoris strain. Here, we recombinantly produced hBD-2 using P. pastoris X-33 as a host. In general, the hBD-2 we produced can be used for therapeutic purposes, and its effectiveness can be evaluated with different experimental studies. Although the in vivo activity of the material was not evaluated in this study, the in vitro antibacterial activity of hBD-2 was investigated. Thus, we confirmed that hBD-2 is actively and recombinantly produced in P. pastoris X-33. Conclusively, it is evident that P. pastoris is an ideal host for the active production of hBD-2, and this system may be used for the further scale-up production of AMPs.
The innate immune system plays an important role in protecting the organism against pathogens as well as initiating inflammatory responses. Since AMP molecules are also produced by the immune system, they form an important component of immunity. AMPs induce numerous immunomodulatory properties by inducing cytokine production, chemoattraction, and immune cell differentiation. This way, they bridge innate immunity to adaptive immunity [44].
Nowadays, infectious diseases are spreading rapidly. As a matter of fact, this has been clearly seen with the COVID-19 pandemic, which has affected the whole world [16]. Since the SARS-CoV-2 virus started to spread, scientists have started to carry out several studies to prevent and treat the disease, and there are still many studies being carried out. In this context, in addition to vaccine studies, the reuse of existing drugs and the screening of possible new drug molecules with in silico approaches are being carried out [16, 45]. [12, 32]. Drugs such as remdesivir, ivermectin, nelfinavir, cepharanthine, hydroxychloroquine, and dexamethasone used for different diseases have been used in clinical trials against SARS-CoV-2[16].
The human ACE-2 receptor is expressed in alveolar epithelial cells. It has many physiological functions in the lungs, gut, kidneys, and brain. ACE-2 has a potential role in treating hypertension and cardiac dysfunction [46]. Furthermore, ACE-2 is vital in innate immunity and for AMP production in the intestines. In COVID-19, ACE-2 is known as a gate through which the virus enters the cell. Therefore, blocking the ACE-2 receptor is an important strategy against the SARS-CoV-2 virus. Another strategy is to inhibit the S protein that interacts with the ACE-2 receptor. AMP molecules interacting with the S protein also have the potential to be used as an antiviral agent against COVID-19.
As a matter of fact, we see one of the examples of this in a study against COVID-19. Human alpha defensin 5 (HD-5), which is a type of enteric defensin, is essential to intestinal immunity. In this study, it was shown that HD-5 binds to the ACE-2 receptor with high affinity. Additionally, HD-5 was reported to reduce the SARS-CoV-2/ACE-2 interaction [47]. Furthermore, it is suggested that COVID-19 is more severe in cases where the HD-5 protein is underproduced and in people with intestinal diseases [47]. Liscano et al. [48] investigated 15 AMP molecules with antiviral properties and their interactions with ACE-2 and the S protein under in silico conditions. They suggested that 2 AMP molecules, Caerin 1.6 and Caerin 1.10, may have potential antiviral properties against COVID-19 through their interaction with the S protein.
On the other hand, the expression of hBD-2 is inducible in response to various infectious agents. The hBD-2 expression in leukocytes shows that it is significant for the adaptive immune system. Idris et al. (2020) [49]. showed that the expression levels of defensin genes were down-regulated in patients with COVID-19. For instance, many defensin genes, such as DEFB4A, 107B, 106B, 4B, and 103 A, are down-regulated in COVID-19 patients. Additionally, copy number variations of defensins also affect the immune system and infections. Moreover, the expression of defensin genes, including hBD-2, is decreased in individuals with weakened immune systems [49].
Xu et al. reported that there was no interaction between hBD-2 and the S protein [50]. However, according to a very recently published study, the hBD-2 protein interacts with the S protein and is among the AMPs that are effective against COVID-19 [51]. Its interaction with the S protein is suggested to occur through its binding to the ACE-2 binding site. The reason for the difference between these two studies may be due to the production of the hBD-2 protein by chemical synthesis and its different isoforms from recombinant production using the E. coli host [51]. These studies include what the host is in recombinant production or whether the synthesis is performed by a chemical method, since these parameters change the properties of the protein that is produced, and this is a situation that must be taken into consideration. In our study, the interaction between hBD-2 and ACE-2 was investigated by both in silico and in vitro analyses. Therefore, ACE-2-hBD-2 binding in our study may also have resulted from our recombinant production of hBD-2 in P. pastoris. This is because post-translational modifications during recombinant production are significant and may vary depending on the recombinant production host.
According to our results, it appears that the hBD-2 protein produced in P. pastoris made post-translational modifications to interact with the ACE-2 receptor. Here, we may state that hBD-2 produced recombinantly in P. pastoris interacts with and binds to ACE-2. However, our findings need to be supported by cell culture studies, toxicity analyses, and in vivo experiments.
In conclusion, the results we obtained in our study showed that hBD-2 could be produced recombinantly in P. pastoris, and it had functional activity. Additionally, this protein showed a tendency to bind with the ACE-2 receptor protein under both in silico and in vitro conditions.
References
Moretta A, Salvia R, Scieuzo C et al (2020) A bioinformatic study of antimicrobial peptides identified in the black soldier fly (BSF) Hermetia illucens (Diptera: Stratiomyidae). Sci Rep. https://doi.org/10.1038/s41598-020-74017-9
Drayton M, Deisinger JP, Ludwig KC et al (2021) Host defense peptides: dual antimicrobial and immunomodulatory action. Int J Mol Sci
Xu D, Lu W (2020) Defensins: A Double-Edged Sword in Host Immunity. Front Immunol
Ganz T (2003) Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol
Pal M, Berhanu G, Desalegn C, Kandi V (2020) Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2): an update. Cureus. https://doi.org/10.7759/cureus.7423
Sachs JD, Karim SSA, Aknin L et al (2022) The Lancet Commission on lessons for the future from the COVID-19 pandemic. The Lancet
Li F (2016) Structure, function, and evolution of Coronavirus Spike Proteins. Annu Rev Virol
V’kovski P, Kratzel A, Steiner S et al (2021) Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol
Saxena SK, Kumar S, Baxi P et al (2020) Chasing COVID-19 through SARS-CoV-2 spike glycoprotein. Virusdisease
Yıldırım ÖT (2020) COVID-19 pandemisi, anjiyotensin dönüştürücü wnim (ACE) inhibitörleri ve anjiyotensin reseptör blokörlerinin (ARB) kullanımı. Çukurova Anestezi ve Cerrahi Bilimler Dergisi 3:47–52
Hamming I, Cooper ME, Haagmans BL et al (2007) The emerging role of ACE2 in physiology and disease. Journal of Pathology
Harder J, Bartels J, Christophers E, Schroder JM (1997) A peptide antibiotic from human skin [6]. Nature
Schröder JM, Harder J (1999) Human beta-defensin-2. Int J Biochem Cell Biology. https://doi.org/10.1016/S1357-2725(99)00013-8
Zhang L, Ghosh SK, Basavarajappa SC et al (2022) HBD-2 binds SARS-CoV-2 RBD and blocks viral entry: strategy to combat COVID-19. iScience. https://doi.org/10.1016/j.isci.2022.103856
Zhang L, Ghosh SK, Basavarajappa SC et al (2021) Molecular dynamics simulations and functional studies reveal that hBD-2 binds SARS-CoV-2 spike RBD and blocks viral entry into ACE2 expressing cells. bioRxiv. https://doi.org/10.1101/2021.01.07.425621
Rani P, Kapoor B, Gulati M et al (2022) Antimicrobial peptides: a plausible approach for COVID-19 treatment. Expert Opin Drug Discov
Huan Y, Kong Q, Mou H, Yi H (2020) Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front Microbiol
Cao J, de La Fuente-Nunez C, Ou RW et al (2018) Yeast-based Synthetic Biology platform for antimicrobial peptide production. ACS Synth Biol. https://doi.org/10.1021/acssynbio.7b00396
Yazici A, Örtücü S, Taşkin M (2021) Screening and characterization of a novel Antibiofilm polypeptide derived from filamentous Fungi. J Proteom. https://doi.org/10.1016/j.jprot.2020.104075
Green MR, Sambrook J (2018) The basic polymerase chain reaction (PCR). Cold Spring Harb Protoc. https://doi.org/10.1101/pdb.prot095117
Skosyrev VS, Kulesskiy EA, Yakhnin AV et al (2003) Expression of the recombinant antibacterial peptide sarcotoxin IA in Escherichia coli cells. Protein Expr Purif. https://doi.org/10.1016/S1046-5928(02)00697-6
Fu P, Wu J, Gao S et al (2015) The recombinant expression and activity detection of MAF-1 fusion protein. Sci Rep. https://doi.org/10.1038/srep14716
Li W, Tao Y, Song CF et al (2021) Multiple copies of the Fusion Gene cflyC-mzfDB3 enhance the expression of a hybrid antimicrobial peptide in Pichia pastoris. Appl Biochem Microbiol. https://doi.org/10.1134/S0003683821020083
Brunelle JL, Green R (2014) One-dimensional SDS-polyacrylamide gel electrophoresis (1D SDS-PAGE). In: Methods in Enzymology
Shi N, Jin F, Zhang X et al (2014) Overexpression of human β-defensin 2 promotes growth and invasion during esophageal carcinogenesis. Oncotarget. https://doi.org/10.18632/oncotarget.2416
Khatri NK, Hoffmann F (2006) Impact of methanol concentration on secreted protein production in oxygen-limited cultures of recombinant Pichia pastoris. Biotechnol Bioeng. https://doi.org/10.1002/bit.20773
Zavec D, Gasser B, Mattanovich D (2020) Characterization of methanol utilization negative Pichia pastoris for secreted protein production: new cultivation strategies for current and future applications. Biotechnol Bioeng. https://doi.org/10.1002/bit.27303
Cao J, de La Fuente-Nunez C, Ou RW et al (2018) Yeast-based Synthetic Biology platform for antimicrobial peptide production. ACS Synth Biol. https://doi.org/10.1021/acssynbio.7b00396
de Sá Magalhães S, Keshavarz-Moore E (2021) P. Pastoris (komagataella phaffii) as a cost‐effective tool for vaccine production for low‐ and middle‐income countries (lmics). Bioengineering
Yang Z, Zhang Z (2018) Engineering strategies for enhanced production of protein and bio-products in Pichia pastoris: a review. Biotechnol Adv
Meng DM, Zhao JF, Ling X et al (2017) Recombinant expression, purification and antimicrobial activity of a novel antimicrobial peptide PaDef in Pichia pastoris. Protein Expr Purif. https://doi.org/10.1016/j.pep.2016.10.003
Huynh E, Akhtar N, Li J (2018) Efficient production of recombinant Protegrin-1 from Pichia pastoris, and its antimicrobial and in vitro cell Migration Activity. Front Microbiol. https://doi.org/10.3389/fmicb.2018.02300
Mulder KC, de Lima LA, Aguiar PS et al (2015) Production of a modified peptide clavanin in Pichia pastoris: cloning, expression, purification and in vitro activities. AMB Express. https://doi.org/10.1186/s13568-015-0129-0
Peschel A, Sahl HG (2006) The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat Rev Microbiol
Patricia Rosete Olvera D, Cabello Gutiérrez C (2019) Multifunctional activity of the β-Defensin-2 during respiratory infections. In: Immune Response Activation and Immunomodulation
Corrales-García LL, Serrano-Carreón L, Corzo G (2020) Improving the heterologous expression of human β-defensin 2 (HBD2) using an experimental design. Protein Expr Purif. https://doi.org/10.1016/j.pep.2019.105539
Brautaset T, Lale R, Valla S (2009) Positively regulated bacterial expression systems. Microb Biotechnol
Fang X, Peng L, Xu Z et al (2005) Cloning and expression of human Beta-defensin-2 gene in Escherichia Coli. Protein Pept Lett. https://doi.org/10.2174/0929866023409011
Peng L, Xu Z, Fang X et al (2004) High-level expression of soluble human β-defensin-2 in Escherichia coli. Process Biochem. https://doi.org/10.1016/j.procbio.2003.11.011
Xu Z, Peng L, Zhong Z et al (2006) High-level expression of a soluble functional antimicrobial peptide, human β-defensin 2, in Escherichia coli. Biotechnol Prog. https://doi.org/10.1021/bp0502680
Aerts AM, Thevissen K, Bresseleers SM et al (2007) Arabidopsis thaliana plants expressing human beta-defensin-2 are more resistant to fungal attack: functional homology between plant and human defensins. Plant Cell Rep. https://doi.org/10.1007/s00299-007-0329-4
Møller TSB, Hay J, Saxton MJ et al (2017) Human β-defensin-2 production from S. cerevisiae using the repressible MET17 promoter. Microb Cell Fact. https://doi.org/10.1186/s12934-017-0627-7
Bindra GK, Williams SA, Lay FT et al (2022) Human β-Defensin 2 (HBD-2) displays oncolytic activity but does not affect Tumour Cell Migration. Biomolecules. https://doi.org/10.3390/biom12020264
Pahar B, Madonna S, Das A et al (2020) Immunomodulatory role of the antimicrobial ll-37 peptide in autoimmune diseases and viral infections. Vaccines (Basel
Güler H, Ay Şal F, Can Z et al (2021) Targeting cov-2 spike rbd and ace-2 interaction with flavonoids of anatolian propolis by in silico and in vitro studies in terms of possible covid-19 therapeutics. Turkish J Biology. https://doi.org/10.3906/biy-2104-5
Bosso M, Thanaraj TA, Abu-Farha M et al (2020) The two faces of ACE2: the role of ACE2 receptor and its polymorphisms in hypertension and COVID-19. Mol Ther Methods Clin Dev
Wang C, Wang S, Li D et al (2020) Human intestinal defensin 5 inhibits SARS-CoV-2 Invasion by cloaking ACE2. Gastroenterology. https://doi.org/10.1053/j.gastro.2020.05.015
Liscano Y, Oñate-Garzón J, Ocampo-Ibáñez ID (2020) In silico discovery of antimicrobial peptides as an alternative to control sars-cov-2. Molecules. https://doi.org/10.3390/molecules25235535
Idris MM, Banu S, Siva AB, Nagaraj R (2020) Downregulation of Defensin genes in SARS-CoV-2 infection. medRxiv
Xu C, Wang A, Marin M et al (2021) Human defensins inhibit SARS-CoV-2 infection by blocking viral entry. Viruses. https://doi.org/10.3390/v13071246
Zhang L, Ghosh SK, Basavarajappa SC et al (2022) HBD-2 binds SARS-CoV-2 RBD and blocks viral entry: strategy to combat COVID-19. iScience. https://doi.org/10.1016/j.isci.2022.103856
Acknowledgements
This study was supported by The Scientific and Technological Research Council of Turkey (TÜBİTAK) with Project number: 121Z469.
Author information
Authors and Affiliations
Contributions
ŞÇ, EA, AY, and SÖ performed the experiments related to production of hBD-2. This study is a part of ŞÇ’s master thesis. SÖ performed the data interpretation and editing the manuscript and AY carried out the conceptualization, experimental designing, data analysis, writing, and editing of the manuscript. The authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical Approval
Not required.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Çobanoğlu, Ş., Arslan, E., Yazıcı, A. et al. Expression of Human β-defensin 2 (hBD-2) in Pichia Pastoris and Investigation of Its Binding Efficiency with ACE-2. Protein J 42, 399–407 (2023). https://doi.org/10.1007/s10930-023-10130-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-023-10130-8