Abstract
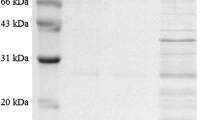
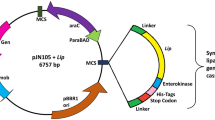
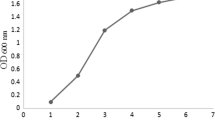
Pfu DNA polymerase is one of the most preferred molecular enzymes that is isolated from the hyperthermophilic Pyrococcus furiosus and used for high-throughput DNA synthesis by the polymerase chain reaction. Therefore, an efficient Pfu DNA polymerase production method is necessary for molecular techniques. In the present study, Pfu DNA polymerase was expressed in recombinant Escherichia coli BL21(DE3) and significant parameters for the biomass production were optimized using the central composite design which is the most popular method of response surface methodology. Induction conditions including cell density prior induction (OD600nm), post-induction temperature, IPTG concentration, and post-induction time and their interactions on biomass production were investigated. The maximum biomass production (14.1 g/L) in shake flasks was achieved using the following predicted optimal conditions: OD600nm before induction of 0.4 and the induction at 32 °C for 7.7 h, with 0.6 mM IPTG. Optimized culture conditions were implemented to scale up experiments. 22% and 70% increase in biomass production was achieved in 3 L and 10 L bioreactors, respectively as compared to initial biomass production observed in unoptimized conditions. Similary, a 30% increase of Pfu DNA polymerase production was obtained after the optimization. The polymerase activity of the purifed Pfu DNA polymerase was assessed by PCR amplification and determined as 2.9 U/μl by comparison with commercial Pfu DNA polymerase. The findings of this study indicated that the proposed fermentation conditions will contribute to further scale‑up studies to enhance the biomass for the production of other recombinant proteins.



Similar content being viewed by others
Data Availability
All data generated or analyzed during this study are included in this published article.
References
Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA (1988) Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239(4839):487–491
Pavlov AR, Pavlova NV, Kozyavkin SA, Slesarev AI (2004) Recent developments in the optimization of thermostable DNA polymerases for efficient applications. Trends Biotechnol 22(5):253–260
Lipps G, Röther S, Hart C, Krauss G (2003) A novel type of replicative enzyme harbouring ATPase, primase and DNA polymerase activity. EMBO J 22(10):2516–2525
Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, Prakash L, Prakash S, Todo T, Walker GC, Wang Z, Woodgate R (2001) The Y-family of DNA polymerases. Mol Cell 8(1):7–8
Joyce CM, Steitz TA (1994) Function and structure relationships in DNA polymerases. Annu Rev Biochem 63:777–822
Ishino S, Ishino Y (2014) DNA polymerases as useful reagents for biotechnology - the history of developmental research in the field. Front Microbiol 5:465
Hansen CJ, Wu L, Fox JD, Arezi B, Hogrefe HH (2011) Engineered split in Pfu DNA polymerase fingers domain improves incorporation of nucleotide gamma-phosphate derivative. Nucleic Acids Res 39(5):1801–1810
Melissis S, Labrou NE, Clonis YD (2006) Nucleotide-mimetic synthetic ligands for DNA-recognizing enzymes—one-step purification of Pfu DNA polymerase. J Chromatogr A 1122(1–2):63–75
Norholm MH (2010) A mutant Pfu DNA polymerase designed for advanced uracil-excision DNA engineering. BMC Biotech 10:21
Cline J, Braman JC, Hogrefe HH (1996) PCR fidelity of Pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res 24(18):3546–3551
McInerney P, Adams P, Hadi MZ (2014) Error rate comparison during polymerase chain reaction by DNA polymerase. Mol Biol Int 2014:287430
Flaman JM, Frebourg T, Moreau V, Charbonnier F, Martin C, Ishioka C, Friend SH, Iggo R (1994) A rapid PCR fidelity assay. Nucleic Acids Res 22(15):3259–3260
Lundberg KS, Shoemaker DD, Adams MWW, Short JM, Sorge JA, Mathur EJ (1991) High-fidelity amplification using a thermostable DNA polymerase isolated from Pyrococcus furiosus. Gene 108(1):1–6
Barnes WM (1994) PCR amplification of up to 35-kb DNA with high fidelity and high yield from lambda bacteriophage templates. PNAS 91(6):2216–2220
Uemori T, Sato Y, Kato I, Doi H, Ishino Y (1997) A novel DNA polymerase in the hyperthermophilic archaeon, Pyrococcus furiosus: gene cloning, expression, and characterization. Genes Cells 2(8):499–512
Uemori T, Ishino Y, Toh H, Asada K, Kato I (1993) Organization and nucleotide sequence of the DNA polymerase gene from the archaeon Pyrococcus furiosus. Nucleic Acids Res 21(2):259–265
Zheng W, Wang Q, Bi Q (2016) Construction, expression and characterization of recombinant Pfu DNA polymerase in Escherichia coli. Protein J 35(2):145–153
Mroczkowski BS, Huvar A, Lernhardt W, Misono K, Nielson K, Scott B (1994) Secretion of thermostable DNA polymerase using a novel baculovirus vector. J Biol Chem 269(18):13522–13528
Lu CL, Erickson HP (1997) Expression in Escherichia coli of the thermostable DNA polymerase from Pyrococcus furiosus. Protein Expr Purif 11(2):179–184
Dabrowski S, Kur J (1998) Cloning and expression in Escherichia coli of the recombinant His-Tagged DNA polymerases from Pyrococcus furiosus and Pyrococc woesei. Protein Expr Purif 14(1):131–138
Mathur EJ (2002) US patent 6:489150
Graslund S, Sagemark J, Berglund H, Dahlgren LG, Flores A, Hammarstroem M, Johansson I, Kotenyova T, Nilsson M, Nordlund P, Weigelt J (2008) The use of systematic N- and C-terminal deletions to promote production and structural studies of recombinant proteins. Protein Expr Purif 58(2):210–221
Mandenius CF, Brundin A (2008) Bioprocess optimization using design of experiments methodology. Biotechnol Prog 24(6):1191–1203
Gilman J, Walls L, Bandiera L, Menolascina F (2021) Statistical design of experiments for synthetic biology. ACS Synth Biol 10(1):1–18
Crater JS, Lievense JC (2018) Scale-up of industrial microbial processes. FEMS Microbiol Lett 365(13):1–5
Evans SJ, Fogg MJ, Mamone A, Davis M, Pearl LH, Connolly BA (2000) Improving dideoxynucleotide-triphosphate utilisation by the hyper-thermophilic DNA polymerase from the archaeon Pyrococcus furiosus. Nucleic Acids Res 28(5):1059–1066
Ceylan HK, Ulusu Y, Bilgin S, Gökçe İ (2021) Expression of cellulose-degrading endoglucanase from Bacillus subtilis using pTolT expression system in Escherichia coli. Cellulose Chem Technol 55(5–6):619–627
Samman N, Al-Muhalhil K, Nehdi A (2023) A simple and efficient method for Taq DNA polymerase purification based on heat denaturation and affinity chromatography. J King Saud Univ- Sci 35(3):102565
Sankar PS, Citartan M, Siti AA, Skryabin BV, Rozhdestvensky TS, Khor GH, Tang TH (2019) A simple method for in-house Pfu DNA polymerase purification for high-fidelity PCR amplification. Iran J Microbiol 11(2):181–186
Teng XC, Nag SY, Citartan M, Tang TH, Ahmed SA (2023) Simple approach for expression and rapid purification of Taq DNA polymerase in three Escherichia coli strains. AsPac J Mol Biol Biotechnol 31(1):45–52
Ceylan HK, Tayhan SE, Gökçe İ (2021) Secretory expression of human vascular endothelial growth factor (VEGF165) in Kluyveromyces lactis and characterization of its biological activity. Int J Pept Res Ther 27:1989–2001
Bozkurt Y, Bilgin S, Erden S, Turan İF, Gökçe İ (2019) Recombinant human G-CSF production as a protein based drug candidate for hematology and oncology. Int J Chem Technol 3(2):92–100
Nemli S, Kianoosh T, Tanyolaç MB (2015) Genetic diversity and population structure of common bean (Phaseolus vulgaris L.) accessions through retrotransposon-based interprimer binding sites (iPBSs) markers. Turk J Agric For 39:940–948
Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak V, Cardona A (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7):676–682
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227(5259):680–685
Vincentelli R, Cimino A, Geerlof A, Kubo A, Satou Y, Cambillau C (2011) Highthroughput protein expression screening and purification in Escherichia coli. Methods 55(1):65–72
Porath J (1992) Immobilized metal-ion affinity-chromatography. Protein Expr Purif 3(4):263–281
Uemori T, Sato Y, Kato I, Doi H, Ishino Y (1997) A novel DNA polymerase in hyperthermophilic archaeon, Pyrococcus furiosus: gene cloning, expression, and characterization. Genes Cells 2(8):499–512
UemoriT IY, Toh H, Asada K, Kato I (1993) Organization and nucleotide sequence of the DNA polymerase gene from the archaeon Pyrococcus furiosus. Nucleic Acids Res 21(2):259–265
Correa A, Oppezzo P (2011) Tuning different expression parameters to achieve soluble recombinant proteins in E. coli: advantages of high-throughput screening. Biotechnol J 6(6):715–730
Gräslund S, Nordlund P, Weigelt J, Hallberg BM, Bray J, Gileadi O, Knapp S, Oppermann U, Arrowsmith C, Hui R, Ming J, Dhe-Paganon S, Park H, Savchenko A, Yee A, Edwards A, Vincentelli R, Cambillau C, Kim R, Kim SH, Rao Z, Shi Y, Terwilliger TC, Kim CY, Hung LW, Waldo GS, Peleg Y, Albeck S, Unger T, Dym O, Prilusky J, Sussman JL, Stevens RC, Lesley SA, Wilson IA, Joachimiak A, Collart F, Dementieva I, Donnelly MI, Eschenfeldt WH, Kim Y, Stols L, Wu R, Zhou M, Burley SK, Emtage JS, Sauder JM, Thompson D, Bain K, Luz J, Gheyi T, Zhang F, Atwell S, Almo SC, Bonanno JB, Fiser A, Swaminathan S, Studier FW, Chance MR, Sali A, Acton TB, Xiao R, Zhao L, Ma LC, Hunt JF, Tong L, Cunningham K, Inouye M, Anderson S, Janjua H, Shastry R, Ho CK, Wang D, Wang H, Jiang M, Montelione GT, Stuart DI, Owens RJ, Daenke S, Schütz A, Heinemann U, Yokoyama S, Büssow K, Gunsalus KC (2008) Protein production and purification. Nat Methods 5(2):135–146
Shijin W, Xiang Y, Zhihang H, Zhang L, Jianmeng C (2009) Optimizing aerobic biodegradation of dichloromethane using response surface methodology. J Environ Sci 21:1276–1283
Singh R, Sooch BS, Puri M (2007) Optimization of medium and process parameters for the production of inulinase from a newly isolated Kluyveromyces marxianus YS-1. Biores Technol 98(13):2518–2525
Maiorano AE, da Silva ES, Perna RF, Ottoni CA, Piccoli RAM, Fernandez RC, Maresma BG, de Andrade Rodrigues MF (2020) Effect of agitation speed and aeration rate on fructosyltransferase production of Aspergillus oryzae IPT-301 in stirred tank bioreactor. Biotechnol Lett 42(12):2619–2629
Pandey R, Kumar N, Prabhu AA, Veeranki VD (2018) Application of medium optimization tools for improving recombinant human interferon gamma production from Kluyveromyces lactis. Prep Biochem Biotechnol 48(3):279–287
Papaneophytou CP, Rinotas V, Douni E, Kontopidis G (2013) A statistical approach for optimization of RANKL overexpression in Escherichia coli: purification and characterization of the protein. Protein Expr Purif 90(1):9–19
Papaneophytou CP, Kontopidis GA (2012) Optimization of TNF-α overexpression in Escherichia coli using response surface methodology: purification of the protein and oligomerization studies. Protein Expr Purif 86(1):35–44
Acknowledgements
This work is supported by the Scientific Research Project Fund of Tokat Gaziosmanpaşa University under project number 2019/39. The author is grateful to Professor Bernard A. Connolly (University of Newcastle, UK) and Professor İsa Gökçe (Tokat Gaziosmanpaşa University, Turkey) for supplying The pET-17b(pfu-Pol) recombinant plasmid. The author thanks Professor Murat Elibol (Ege University, Turkey) for assistance with response surface methodology, Research Assistant Ahmet Düzel and Master of Science Cafer Meydan (Ege University, Turkey) for assistance with the bioreactor experiments, Dr. Rizvan İmamoğlu (Bartın University, Turkey) for supplying the strain E. coli BL21(DE3), Professor Muhammed Bahattin Tanyolaç for providing the necessary laboratory equipment for the activity experiments.
Author information
Authors and Affiliations
Contributions
HKC performed the experiments, analyzed the data, wrote the main manuscript, prepared its figures and revised the manuscript. reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The author declares that he has no conflict of interest.
Ethical Approval
Neither ethical approval nor informed consent was required for this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ceylan, H.K. Enhanced Biomass Production of Recombinant Pfu DNA Polymerase Producer Escherichia coli BL21(DE3) by Optimization of Induction Variables Using Response Surface Methodology. Protein J 42, 451–462 (2023). https://doi.org/10.1007/s10930-023-10122-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-023-10122-8