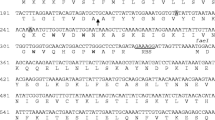
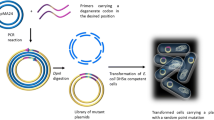
Abstract
To date, there have been no or just a few reports of successful cloning and expression to create biologically active ocins or bacteriocins. Cloning, expression, and production of class I ocins are problematic because of their structural arrangements, coordinated functions, size, and posttranslational modifications. Mass synthesis of these molecules is necessary for commercialization and to restrict the excessive use of conventional antibiotics, which encourages the development of antibiotic-resistant bacteria. In the case of class III ocins, there are no reports of obtaining biological active proteins to date. Being able to obtain biologically active proteins requires an understanding of mechanistic features due to their expanding importance and broad spectrum of activity. As a result, we intend to clone and express the class III type. The class I types that are devoid of posttranslational modifications were transformed into class III through fusion. Therefore, this construct resembles a class III type ocin. With the exception of Zoocin, expression of the proteins was found to be physiologically ineffective after cloning. But, few cell morphological changes such as elongation, aggregation, and the formation of terminal hyphae were observed. However, it was discovered that the target indicator had been altered to Vibrio spp. in a few. All the three ocins were subjected to in-silico structure prediction/analysis. Finally, we confirm the existence of unidentified additional intrinsic factors for successful expression to obtain biologically active protein.



Similar content being viewed by others
References
Mokoena MP (2017) Lactic acid bacteria and their bacteriocins: classification, biosynthesis and applications against uropathogens: a mini-review. Molecules 22(8):1255. https://doi.org/10.3390/molecules22081255
Simons A, Alhanout K, Duval RE (2020) Bacteriocins, antimicrobial peptides from bacterial origin: overview of their biology and their impact against multidrug-resistant bacteria. Microorganisms 8(5):639. https://doi.org/10.3390/microorganisms8050639
Tajbakhsh M, Karimi A, Fallah F, Akhavan MM (2017) Overview of ribosomal and non-ribosomal antimicrobial peptides produced by Gram positive bacteria. Cell Mol Biol (Noisy-le-Grand) 63(10):20–32. https://doi.org/10.14715/cmb/2017.63.10.4
Ingham AB, Sproat KW, Tizard ML et al (2005) A versatile system for the expression of non-modified bacteriocins in Escherichia coli. J Appl Microbiol 98(3):676–683. https://doi.org/10.1111/j.1365-2672.2004.02502.x
Alvarez-Sieiro P, Montalban-Lopez M, Mu D et al (2016) Bacteriocins of lactic acid bacteria: extending the family. Appl Microbiol Biotechnol 100(7):2939–2951. https://doi.org/10.1007/s00253-016-7343-9
Yi L, Luo L, Lu X (2018) Efficient exploitation of multiple novel bacteriocins by combination of complete genome and peptidome. Front Microbiol 9:1567. https://doi.org/10.3389/fmicb.2018.01567
Kumaria R, Garsa AK, Rajput YS et al (2019) Bacteriocins: classification, synthesis, mechanism of action and resistance development in food spoilage causing bacteria. Microb Pathog 128:171–177. https://doi.org/10.1016/j.micpath.2019.01.002
Abee T, Krockel L, Hill C (1995) Bacteriocins: modes of action and potentials in food preservation and control of food poisoning. Int J Food Microbiol 28(2):169–185. https://doi.org/10.1016/0168-1605(95)00055-0
O’Connor PM, O’Shea EF, Cotter PD et al (2018) The potency of the broad spectrum bacteriocin, bactofencin A, against staphylococci is highly dependent on primary structure, N-terminal charge and disulphide formation. Sci Rep 8(1):1. https://www.researchgate.net/publication/326879573
Xin J et al (2019) Heterologous expression and purification of BtCsPB, a Novel Cold-shock protein like bacteriocin from Bacillus thurungenesis BRC-ZYR2. World J Microbiol Biotech 35(2):1–5. https://doi.org/10.1007/s11274-019-2595-z
Wibowo D, Zhao CX (2019) Recent achievements and perspectives for large-scale recombinant production of antimicrobial peptides. Appl Microbiol Biotechnol 103(2):659–671. https://doi.org/10.1007/s00253-018-9524-1
Gopal GJ, Kumar A (2013) Strategies for the production of recombinant protein in Escherichia coli. Protein J 32:419–425
Klocke M, Mundt K, Idler F et al (2005) Heterologous expression of enterocin A, a bacteriocin from Enterococcus faecium, fused to a cellulose-binding domain in Escherichia coli results in a functional protein with inhibitory activity against Listeria. Appl Microbiol Biotechnol 67(4):532–538. https://doi.org/10.1007/s00253-004-1838-5
Choyam S, Suresh PSN, Pandey R et al (2019) Ocins database: a database of bug-busters from Bifidobacterium, Lactobacillus, and Enterococcus. Access Microbiol 1:4
Hammami R, Zouhir A, Le Lay C et al (2010) BACTIBASE second release: a database and tool platform for bacteriocin characterization. BMC Microbiol 10:22
Mesa-Pereira B, Rea MC, Cotter PD et al (2018) Heterologous expression of bio-preservative bacteriocins with a view to low cost production. Front Microbiol. https://doi.org/10.3389/fmicb.2018.01654
Schreiber C, Muller H, Birrenbach O et al (2017) A high-throughput expression screening platform to optimize the production of antimicrobial peptides. Microb Cell Fact 16(1):29. https://doi.org/10.1186/s12934-017-0637-5
Gibbs GM, Davidson BE, Hillier AJ (2004) Novel expression system for large-scale production and purification of recombinant class IIa bacteriocins and its application to piscicolin 126. Appl Environ Microbiol 70(6):3292–3297. https://doi.org/10.1128/AEM.70.6.3292-3297.2004
Acuna L, Picariello G, Sesma F et al (2012) A new hybrid bacteriocin, Ent35-MccV, displays antimicrobial activity against pathogenic Gram-positive and Gram-negative bacteria. FEBS Open Bio 2:12–19. https://doi.org/10.1016/j.fob.2012.01.002
Sal-Man N, Oren Z, Shai Y (2002) Preassembly of membrane-active peptides is an important factor in their selectivity toward target cells. Biochemistry 41(39):11921–11930. https://doi.org/10.1021/bi0260482
Arai R, Ueda H, Kitayama A et al (2001) Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng 14(8):529–532. https://doi.org/10.1093/protein/14.8.529
Klein JS, Jiang S, Galimidi RP et al (2014) Design and characterization of structured protein linkers with differing flexibilities. Protein Eng Des Sel 27(10):325–330. https://doi.org/10.1093/protein/gzu043
Reddy Chichili VP, Kumar V, Sivaraman J (2013) Linkers in the structural biology of protein–protein interactions. Protein Sci 22(2):153–167. https://doi.org/10.1002/pro.2206
He J, Eckert R, Pharm T et al (2007) Novel synthetic antimicrobial peptides against Streptococcus mutans. Antimicrob Agents Chemother 51(4):1351–1358. https://doi.org/10.1128/AAC.01270-06
Baltzar BK, Jorgenson MG (2017) Recombinant hybrid bacteriocins inhibit the growth of bacterial pathogens. PLoS Synth Biol 2017:1–10
Maarten GK, Kemland GL, Anoz-Carbonell E et al (2017) A natural chimeric pseudomonas bacteriocin with novel pore-forming activity parasitizes the ferrichrome transporter. mBio 8(1):e01961-16. https://doi.org/10.1128/mBio.01961-16
van Rosmalen M, Krom M, Merkx M (2017) Tuning the flexibility of glycine-serine linkers to allow rational design of multi domain proteins. Biochemistry 56(50):6565–6574. https://doi.org/10.1021/acs.biochem.7b00902
Tiwari SK, Sutyak Noll K, Cavera VL et al (2015) Improved antimicrobial activities of synthetic-hybrid bacteriocins designed from enterocin E50–52 and pediocin PA-1. Appl Environ Microbiol 81(5):1661–1667. https://doi.org/10.1128/AEM.03477-14
Acuna L, Morero RD, Bellomio A (2010) Development of wide spectrum hybrid bacteriocins for food bio-preservation. Food Bioprocess Technol 4:1029–1049. https://doi.org/10.1007/s11947-010-0465-7
Guo C, Huang Y, Zheng H et al (2012) Secretion and activity of antimicrobial peptide cecropin D expressed in Pichia pastoris. Exp Ther Med. https://doi.org/10.3892/etm.2012.719
Kageyama M, Kobayashi M, Sano Y (1996) Construction and characterization of pyocin–colicin chimeric proteins. J Bacteriol 178(1):103–110. https://doi.org/10.1128/jb.178.1.103-110.1996
van Rosmalen M, Krom M, Merkx M (2017) Tuning the flexibility of glycine-serine linkers to allow rational design of multi-domain proteins. Biochemistry 56(50):6565–6574. https://doi.org/10.1021/acs.biochem.7b00902
Chen X, Zaro JL, Shen WC (2013) Fusion protein linkers: property, design and functionality. Adv Drug Deliv Rev 65(10):1357–1369. https://doi.org/10.1016/j.addr.2012.09.039
Reddy Chichili VP, Kumar V, Sivaraman J (2013) Linkers in the structural biology of protein–protein interactions. Protein Sci 22(2):153–167. https://doi.org/10.1002/pro.2206
Yi L, Luo L, Lü X (2018) Efficient exploitation of multiple novel bacteriocins by combination of complete genome and peptidome. Front Microbiol 9:1567. https://doi.org/10.3389/fmicb.2018.01567
Valdés-Stauber N, Scherer S (1994) Isolation and characterization of Linocin M18, a bacteriocin produced by Brevibacterium linens. Appl Environ Microbiol 60(10):3809–3814. https://doi.org/10.1128/aem.60.10.3809-3814.1994
Torreblanca M, Meseguer I, Ventosa A (2008) Production of halocin is a practically universal feature of archaeal halophilic rods. Lett Appl Microbiol 19(4):201–205. https://doi.org/10.1111/j.1472-765X.1994.tb00943.x
Mei SS, Li Y, Lu QH et al (2006) Cloning and analysis of genes in the halocin C8 gene cluster. Acta Microbiol Sinica 46(2):318–322
Simmonds RS, Simpson WJ, Tagg JR (1997) Cloning and sequence analysis of zooA, a Streptococcus zoo-epidemicus gene encoding a bacteriocin-like inhibitory substance having a domain structure similar to that of lysostaphin. Gene 189(2):255–261. https://doi.org/10.1016/S0378-1119(96)00859-1
Horton RM, Hunt HD, Ho SN et al (1989) Engineering hybrid genes without the use of restriction enzymes—gene-splicing by overlap extension. Gene 77(1):61–68. https://doi.org/10.1016/0378-1119(89)90359-4
Heckman KL, Pease LR (2007) Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc 2(4):924–932. https://doi.org/10.1038/nprot.2007.132
Thornton JA (2016) Splicing by overlap extension PCR to obtain hybrid DNA products. Methods Mol Biol 1373:43–49. https://doi.org/10.1007/7651_2014_182
Ho SN, Hunt HD, Horton RM et al (1989) Site-directed mutagenesis by overlap extension using the polymerase chain-reaction. Gene 77(1):51–59. https://doi.org/10.1016/0378-1119(89)90358-2
Inoue H, Nojima H, Okayama H (1990) High efficiency transformation of E. coli with plasmids. Gene 96(1):23–28. https://doi.org/10.1016/0378-1119(90) (p. 90336)
Schagger H (2006) Tricine-SDS-PAGE. Nat Protoc 1(1):16–22. https://doi.org/10.1038/nprot.2006.4
Zheng W, Zhang C, Li Y (2021) Folding non-homology proteins by coupling deep-learning contact maps with I-TASSER assembly simulations. Cell Rep Methods 1:100014
Sitao Wu, Zhang Y (2007) LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res 35:3375–3382
LeDuc RD, Fellers RT, Early BP, Thomas PM, Kelleher NL (2014) The C-score: a Bayesian framework to sharply improve proteo-form scoring in high-throughput top down proteomics. J Proteome Res 13(7)3231–3240. https://doi.org/10.1021/pr401277r
Teich A, Lin HY, Andersson L (1998) Amplification of ColE1 related plasmids in recombinant cultures of E. coli after IPTG induction. J of Biotechnol 64:197–210
Vind J, Sørenson MA, Rasmussen MD (1993) Synthesis of proteins in Escherichia coli is limited by the concentration of free ribosomes. Expression from reporter genes does not always reflect functional mRNA levels. J Mol Biol 231:678–688
Fakruddin M, Mazumdar RM, Mannan KSB (2013) Critical factors affecting the success of cloning, expression, and mass production of enzymes by recombinant E. coli. ISRN Biotechnol. https://doi.org/10.5402/2013/590587
Miller Kurt W, Schamber R, Osmanagaoglu O (1998) Isolation and characterization of pediocin AcH chimeric protein mutants with altered bactericidal activity. Appl Env Microbiol 64(6):1997–2005
Sara A, Jimenez JJ, Gutiez L, Feito J (2019) Cloning and expression of synthetic genes encoding native, hybrid- and bacteriocin derived chimeras from mature class IIa bacteriocins, by Pichia pastoris (syn. Komagataella spp). Food Res Int. https://doi.org/10.1016/j.foodres.2019.01.015
Carrier MJ, Nugent ME, Tacon WCA, Primrose SB (1983) High expression of cloned genes in E. coli and its consequences. Trends Biotechnol 14:109-113
Johnsen L, Fimland G, Nissen-Meyer J (2005) The C-terminal domain of pediocin-like antimicrobial peptides (class IIa bacteriocins) is involved in specific recognition of the C-terminal part of cognate immunity proteins and in determining the antimicrobial spectrum. J Bio Chem 280(10): 9243–9250
Fimland G, Blingsmo OR, Sletten K (1996) New biologically active hybrid bacteriocins constructed by combining regions from various pediocin-like bacteriocins: the C-terminal region is important for determining specificity. Appl Environ Microbiol 62(9):3313–3318
Wiedmann I, Breukink E, Van Kraij C et al (2001) Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J Biol Chem 276(3):1772–1779. https://doi.org/10.1074/jbc.M006770200
Morton JT, Freed SD, Lee SW et al (2015) A large-scale prediction of bacteriocin gene blocks suggests a wide functional spectrum for bacteriocins. BMC Bioinform. https://doi.org/10.1186/s12859-015-0792-9
Author information
Authors and Affiliations
Contributions
SC: Generated data, RK: Planned, framed, analysed and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Choyam, S., Kammara, R. Understanding the Necessity of Regulatory Protein Machinery in Heterologous Expression of Class-III Type of Ocins. Protein J 42, 239–252 (2023). https://doi.org/10.1007/s10930-023-10106-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-023-10106-8