Abstract
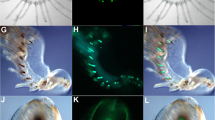
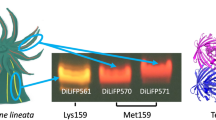
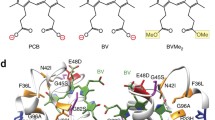
Biofluorescence has been found to be an increasingly widespread phenomenon in the ocean. The reclusive Caribbean chlopsid eel, Kaupichthys hyoproroides displays bright green fluorescence in its native marine environment. We have previously shown the fluorescence to be attributed to a fluorescent fatty acid-binding protein, Chlopsid FP, part of a larger family of fluorescent fatty acid-binding proteins, including the homologous UnaG. All require the addition of exogenous bilirubin for fluorescence. Here, we report the generation of a series of point mutants, and deletions that result in the quenching of fluorescence in Chlopsid FP. In addition, we report the binding constants of bilirubin to Chlopsid FP and mutants, measured by fluorescence titration. This study provides key insights into the potential mechanism of fluorescence in this class of fluorescent fatty acid-binding proteins.





Similar content being viewed by others
References
Sparks JS, Schelly RC, Smith WL et al (2014) The covert world of fish biofluorescence: a phylogenetically widespread and phenotypically variable phenomenon. PLoS ONE 9:e83259. https://doi.org/10.1371/journal.pone.0083259
Gruber DF, Loew ER, Deheyn DD et al (2016) Biofluorescence in catsharks (Scyliorhinidae): fundamental description and relevance for elasmobranch visual ecology. Sci Rep. https://doi.org/10.1038/srep24751
Salih A, Larkum A, Cox G et al (2000) Fluorescent pigments in corals are photoprotective. Nature 408:850. https://doi.org/10.1038/35048564
Rye C, Wise R, Jurukovski V, et al Aquatic biomes: biology 2e. OpenStax
Tyler J (1968) JERLOV, N. G. 1968. Optical oceanography. American Elsevier Publ. Co., Inc, New York. 194 p. $13.50. Limnol Oceanogr 13:731–732. https://doi.org/10.4319/lo.1968.13.4.0731
Gruber DF, Gaffney JP, Mehr S et al (2015) Adaptive evolution of eel fluorescent proteins from fatty acid binding proteins produces bright fluorescence in the marine environment. PLoS ONE. https://doi.org/10.1371/journal.pone.0140972
Kumagai A, Ando R, Miyatake H et al (2013) A bilirubin-inducible fluorescent protein from eel muscle. Cell 153:1602–1611. https://doi.org/10.1016/j.cell.2013.05.038
Shitashima Y, Shimozawa T, Kumagai A et al (2017) Two distinct fluorescence states of the ligand-induced green fluorescent protein UnaG. Biophys J 113:2805–2814. https://doi.org/10.1016/j.bpj.2017.10.022
Shitashima Y, Shimozawa T, Asahi T, Miyawaki A (2018) A dual-ligand-modulable fluorescent protein based on UnaG and calmodulin. Biochem Biophys Res Commun 496:872–879. https://doi.org/10.1016/j.bbrc.2018.01.134
Hayashi S, Toda Y (2009) A novel fluorescent protein purified from eel muscle. Fish Sci 75:1461–1469. https://doi.org/10.1007/s12562-009-0176-z
Funahashi A, Itakura T, Hassanin AAI et al (2017) Ubiquitous distribution of fluorescent protein in muscles of four species and two subspecies of eel (genus Anguilla). J Genet 96:127–133. https://doi.org/10.1007/s12041-017-0751-5
Aoyama J, Nishida M, Tsukamoto K (2001) Molecular phylogeny and evolution of the freshwater eel, genus anguilla. Mol Phylogenet Evol 20:450–459. https://doi.org/10.1006/mpev.2001.0959
Broichhagen J, Trauner D (2013) Bilirubin in a new light. Angew Chem Int Ed 52:13868–13870. https://doi.org/10.1002/anie.201307345
Yu C-L, Ferraro D, Ramaswamy S et al (2008) Purification and properties of Sandercyanin, a blue protein secreted in the mucus of blue forms of walleye, Sander vitreus. Environ Biol Fish 82:51–58. https://doi.org/10.1007/s10641-007-9252-3
Ghosh S, Yu C-L, Ferraro DJ et al (2016) Blue protein with red fluorescence. PNAS 113:11513–11518. https://doi.org/10.1073/pnas.1525622113
Flower DR (1996) The lipocalin protein family: structure and function. Biochem J 318:1–14
Storch J, Thumser AEA (2000) The fatty acid transport function of fatty acid-binding proteins. Biochem Biophys Acta 1486:28–44. https://doi.org/10.1016/S1388-1981(00)00046-9
Zimmerman AW, Veerkamp JH (2002) New insights into the structure and function of fatty acid-binding proteins. Cell Mol Life Sci 59:1096–1116. https://doi.org/10.1007/s00018-002-8490-y
Schaap FG, van der Vusse GJ, Glatz JFC (2002) Evolution of the family of intracellular lipid binding proteins in vertebrates. In: Glatz JFC (ed) Cellular lipid binding proteins. Springer, Boston, pp 69–77
Chmurzyńska A (2006) The multigene family of fatty acid-binding proteins (FABPs): function, structure and polymorphism. J Appl Genet 47:39–48. https://doi.org/10.1007/BF03194597
Hu H, Wang A, Huang L et al (2018) Monitoring cellular redox state under hypoxia using a fluorescent sensor based on eel fluorescent protein. Free Radical Biol Med 120:255–265. https://doi.org/10.1016/j.freeradbiomed.2018.03.041
Adeosun SO, Moore KH, Lang DM, et al. (2018) A novel fluorescence-based assay for the measurement of biliverdin reductase activity. React Oxyg Species 5:35–45. https://doi.org/10.20455/ros.2018.809
Inoué S, Shimomura O, Goda M et al (2002) Fluorescence polarization of green fluorescence protein. Proc Natl Acad Sci USA 99:4272–4277. https://doi.org/10.1073/pnas.062065199
Pollard TD (2010) A guide to simple and informative binding assays. Mol Biol Cell 21:4061–4067. https://doi.org/10.1091/mbc.E10-08-0683
KaleidaGraph. Synergy Software
Hulme EC, Trevethick MA (2010) Ligand binding assays at equilibrium: validation and interpretation. Br J Pharmacol 161:1219–1237. https://doi.org/10.1111/j.1476-5381.2009.00604
The PyMOL Molecular Graphics System. Version 2.0. Schrodinger, LLC
Waterhouse A, Bertoni M, Bienert S et al (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303. https://doi.org/10.1093/nar/gky427
Bienert S, Waterhouse A, de Beer TAP et al (2017) The SWISS-MODEL repository-new features and functionality. Nucleic Acids Res 45:D313–D319. https://doi.org/10.1093/nar/gkw1132
Guex N, Peitsch MC, Schwede T (2009) Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30(Suppl 1):S162–173. https://doi.org/10.1002/elps.200900140
Benkert P, Biasini M, Schwede T (2011) Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27:343–350. https://doi.org/10.1093/bioinformatics/btq662
Bertoni M, Kiefer F, Biasini M et al (2017) Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci Rep. https://doi.org/10.1038/s41598-017-09654-8
Yeh JT-H, Nam K, Yeh JT-H, Perrimon N (2017) eUnaG: a new ligand-inducible fluorescent reporter to detect drug transporter activity in live cells. Sci Rep. https://doi.org/10.1038/srep41619
Acknowledgements
This work was supported by a National Science Foundation Career Grant to J.P.G (Award Number MCB 1652731). We would like to thank Professor Pablo Peixoto for use of his fluorimeter.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Krivoshik, S.R., Guarnaccia, A.M., Fried, D.B. et al. Disrupting Fluorescence by Mutagenesis in a Green Fluorescent Fatty Acid Binding Protein from a Marine Eel. Protein J 39, 145–151 (2020). https://doi.org/10.1007/s10930-020-09883-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-020-09883-3