Abstract
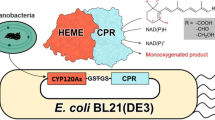
The bioconversion of vitamin D3 catalyzed by cytochrome P450 (CYP) requires 25-hydroxylation and subsequent 1α-hydroxylation to produce the hormonal activated 1α,25-dihydroxyvitamin D3. Vitamin D3 25-hydroxylase catalyses the first step in the vitamin D3 biosynthetic pathway, essential in the de novo activation of vitamin D3. A CYP known as CYP107CB2 has been identified as a novel vitamin D hydroxylase in Bacillus lehensis G1. In order to deepen the understanding of this bacterial origin CYP107CB2, its detailed biological functions as well as biochemical characteristics were defined. CYP107CB2 was characterized through the absorption spectral analysis and accordingly, the enzyme was assayed for vitamin D3 hydroxylation activity. CYP-ligand characterization and catalysis optimization were conducted to increase the turnover of hydroxylated products in an NADPH-regenerating system. Results revealed that the over-expressed CYP107CB2 protein was dominantly cytosolic and the purified fraction showed a protein band at approximately 62 kDa on SDS–PAGE, indicative of CYP107CB2. Spectral analysis indicated that CYP107CB2 protein was properly folded and it was in the active form to catalyze vitamin D3 reaction at C25. HPLC and MS analysis from a reconstituted enzymatic reaction confirmed the hydroxylated products were 25-hydroxyitamin D3 and 1α,25-dihydroxyvitamin D3 when the substrates vitamin D3 and 1α-hydroxyvitamin D3 were used. Biochemical characterization shows that CYP107CB2 performed hydroxylation activity at 25 °C in pH 8 and successfully increased the production of 1α,25-dihydroxyvitamin D3 up to four fold. These findings show that CYP107CB2 has a biologically relevant vitamin D3 25-hydroxylase activity and further suggest the contribution of CYP family to the metabolism of vitamin D3.








Similar content being viewed by others
References
Jin S, Bryson T, Dawson J (2004) Hydroperoxoferric heme intermediate as a second electrophilic oxidant in cytochrome P450-catalyzed reactions. J Biol Inorg Chem 9:644–653. https://doi.org/10.1007/s00775-004-0575-7
Zurek J, Foloppe N, Harvey JN, Mulholland AJ (2006) Mechanisms of reaction in cytochrome P450: hydroxylation of camphor in P450cam. Org Biomol Chem 4:3931–3937. https://doi.org/10.1039/b611653a
Munro AW, Girvan HM, McLean KJ (2007) Variations on a (t)heme–novel mechanisms, redox partners and catalytic functions in the cytochrome P450 superfamily. Nat Prod Rep 24:585–609. https://doi.org/10.1039/b604190f
Schlichting I, Berendzen J, Chu K et al (2000) The catalytic pathway of cytochrome P450cam at atomic resolution. Science 287:1615–1622. https://doi.org/10.1126/science.287.5458.1615
Julsing MK, Cornelissen S, Bühler B, Schmid A (2008) Heme-iron oxygenases: powerful industrial biocatalysts? Curr Opin Chem Biol 12:177–186. https://doi.org/10.1016/j.cbpa.2008.01.029
Urlacher VB, Girhard M (2012) Cytochrome P450 monooxygenases: an update on perspectives for synthetic application. Trends Biotechnol 30:26–36. https://doi.org/10.1016/j.tibtech.2011.06.012
Sakaki T, Sugimoto H, Hayashi K et al (2011) Bioconversion of vitamin D to its active form by bacterial or mammalian cytochrome P450. Biochim Biophys Acta 1814:249–256. https://doi.org/10.1016/j.bbapap.2010.07.014
Nykjaer A, Dragun D, Walther D et al (1999) An endocytic pathway essential for renal uptake and activation of the steroid 25-(OH) vitamin D3. Cell 96:507–515. https://doi.org/10.1016/S0092-8674(00)80655-8
Zhu JG, Ochalek JT, Kaufmann M et al (2013) CYP2R1 is a major, but not exclusive, contributor to 25-hydroxyvitamin D production in vivo. Proc Natl Acad Sci USA 110:15650–15655. https://doi.org/10.1073/pnas.1315006110
Agmon-Levin N, Theodor E, Segal RM, Shoenfeld Y (2013) Vitamin D in systemic and organ-specific autoimmune diseases. Clin Rev Allergy Immunol 45:256–266. https://doi.org/10.1007/s12016-012-8342-y
Garland C, Gorham E, Mohr S, Garland F (2009) Vitamin D for cancer prevention: global perspective. Ann Epidemiol 19:468–483. https://doi.org/10.1016/j.annepidem.2009.03.021
Lavie CJ, Lee JH, Milani RV (2011) Vitamin D and cardiovascular disease will it live up to its hype?. J Am Coll Cardiol 58:1547–1556. https://doi.org/10.1016/j.jacc.2011.07.008
Holick MF (2004) Vitamin D: importance in the prevention of cancers, type 1 diabetes, heart disease, and osteoporosis. Am J Clin Nutr 79:362–371
Zhu J, Deluca HF (2012) Vitamin D 25-hydroxylase—four decades of searching, are we there yet?. Arch Biochem Biophys 523:30–36. https://doi.org/10.1016/j.abb.2012.01.013
Aiba I, Yamasaki T, Shinki T et al (2006) Characterization of rat and human CYP2J enzymes as Vitamin D 25-hydroxylases. Steroids 71:849–856. https://doi.org/10.1016/j.steroids.2006.04.009
Yamasaki T, Izumi S, Ide H, Ohyama Y (2004) Identification of a novel rat microsomal vitamin D3 25-hydroxylase. J Biol Chem 279:22848–22856. https://doi.org/10.1074/jbc.M311346200
Guo YD, Strugnell S, Back DW, Jones G (1993) Transfected human liver cytochrome P-450 hydroxylates vitamin D analogs at different side-chain positions. Proc Natl Acad Sci USA 90:8668–8672
Cheng JB, Motola DL, Mangelsdorf DJ, Russell DW (2003) De-orphanization of cytochrome P450 2R1: a microsomal vitamin D 25-hydroxilase. J Biol Chem 278:38084–38093. https://doi.org/10.1074/jbc.M307028200
Gupta RP, Hollis BW, Patel SB et al (2004) CYP3A4 is a human microsomal vitamin D 25-hydroxylase. J Bone Miner Res 19:680–688. https://doi.org/10.1359/JBMR.0301257
Rahmaniyan M, Patrick K, Bell NH (2005) Characterization of recombinant CYP2C11: a vitamin D 25-hydroxylase and 24-hydroxylase. Am J Physiol Endocrinol Metab 288:E753–E760. https://doi.org/10.1152/ajpendo.00201.2004
Hosseinpour F, Wikvall K (2000) Porcine microsomal vitamin D(3) 25-hydroxylase (CYP2D25). Catalytic properties, tissue distribution, and comparison with human CYP2D6. J Biol Chem 275:34650–34655. https://doi.org/10.1074/jbc.M004185200
Mclean KJ, Girvan HM, Mason AE et al (2011) Structure, mechanism and function of cytochrome P450 enzymes. In: Samuel P, de Visser DK (eds) Iron-containing enzymes: versatile catalysts of hydroxylation reactions. The Royal Society of Chemistry, Cambridge, p 255–280
Werck-Reichhart D, Feyereisen R (2000) Cytochromes P450: a success story. Genome Biol. https://doi.org/10.1186/gb-2000-1-6-reviews3003
Bernhardt R (2006) Cytochromes P450 as versatile biocatalysts. J Biotechnol 124:128–145. https://doi.org/10.1016/j.jbiotec.2006.01.026
Lewis DFV, Wiseman A (2005) A selective review of bacterial forms of cytochrome P450 enzymes. Enzyme Microb Technol 36:377–384. https://doi.org/10.1016/j.enzmictec.2004.07.018
Hilker BL, Fukushige H, Hou C, Hildebrand D (2008) Comparison of Bacillus monooxygenase genes for unique fatty acid production. Prog Lipid Res 47:1–14. https://doi.org/10.1016/j.plipres.2007.09.003
Budde M, Maurer SC, Schmid RD, Urlacher VB (2004) Cloning, expression and characterisation of CYP102A2, a self-sufficient P450 monooxygenase from Bacillus subtilis. Appl Microbiol Biotechnol 66:180–186. https://doi.org/10.1007/s00253-004-1719-y
Urlacher VB, Eiben S (2006) Cytochrome P450 monooxygenases: perspectives for synthetic application. Trends Biotechnol 24:324–330. https://doi.org/10.1016/j.tibtech.2006.05.002
Noor YM, Samsulrizal NH, Jema’on NA et al (2014) A comparative genomic analysis of the alkalitolerant soil bacterium Bacillus lehensis G1. Gene 545:253–261. https://doi.org/10.1016/j.gene.2014.05.012
Ang SS, Salleh AB, Chor ALT et al (2015) Molecular characterization, modeling and docking of CYP107CB2 from Bacillus lehensis G1, an alkaliphile. Comput Biol Chem 56:19–29. https://doi.org/10.1016/j.compbiolchem.2015.02.015
Ryo Omura T, Sato (1964) The carbon monoxide-biding pigment of liver microsomes. J Biol Chem 239:2370–2378
Fujii Y, Kabumoto H, Nishimura K et al (2009) Purification, characterization, and directed evolution study of a vitamin D3 hydroxylase from Pseudonocardia autotrophica. Biochem Biophys Res Commun 385:170–175. https://doi.org/10.1016/j.bbrc.2009.05.033
Nelson DR (2006) Cytochrome P450 nomenclature, 2004. Methods Mol Biol 320:1–10. https://doi.org/10.1385/1-59259-998-2:1
Schallmey A, den Besten G, Teune IGP et al (2011) Characterization of cytochrome P450 monooxygenase CYP154H1 from the thermophilic soil bacterium Thermobifida fusca. Appl Microbiol Biotechnol 89:1475–1485. https://doi.org/10.1007/s00253-010-2965-9
Green AJ, Rivers SL, Cheesman M et al (2001) Expression, purification and characterization of cytochrome P450 Biol: a novel P450 involved in biotin synthesis in Bacillus subtilis. J Biol Inorg Chem 6:523–533. https://doi.org/10.1007/s007750100229
Yasutake Y, Fujii Y, Nishioka T et al (2010) Structural evidence for enhancement of sequential vitamin D3 hydroxylation activities by directed evolution of cytochrome P450 vitamin D3 hydroxylase. J Biol Chem 285:31193–31201. https://doi.org/10.1074/jbc.M110.147009
Prior JE, Shokati T, Christians U, Gill RT (2010) Identification and characterization of a bacterial cytochrome P450 for the metabolism of diclofenac. Appl Microbiol Biotechnol 85:625–633. https://doi.org/10.1007/s00253-009-2135-0
Sugimoto H, Shinkyo R, Hayashi K et al (2008) Crystal structure of CYP105A1 (P450SU-1) in complex with 1alpha,25-dihydroxyvitamin D3. Biochemistry 47:4017–4027. https://doi.org/10.1021/bi7023767
Roberts GA, Çelik A, Hunter DJB et al (2003) A self-sufficient cytochrome P450 with a primary structural organization that includes a flavin domain and a [2Fe-2S] redox center. J Biol Chem 278:48914–48920. https://doi.org/10.1074/jbc.M309630200
Guengerich FP, Martin MV, Sohl CD, Cheng Q (2009) Measurement of cytochrome P450 and NADPH-cytochrome P450 reductase. Nat Protoc 4:1245–1251. https://doi.org/10.1038/nprot.2009.121
Dunford AJ, McLean KJ, Sabri M et al (2007) Rapid P450 heme iron reduction by laser photoexcitation of Mycobacterium tuberculosis CYP121 and CYP51B1. Analysis of CO complexation reactions and reversibility of the P450/P420 equilibrium. J Biol Chem 282:24816–24824. https://doi.org/10.1074/jbc.M702958200
Sasaki M, Akahira A, Oshiman K et al (2005) Purification of cytochrome P450 and ferredoxin, involved in bisphenol A degradation, from Sphingomonas sp. strain AO1. Appl Environ Microbiol 71:8024–8030. https://doi.org/10.1128/AEM.71.12.8024-8030.2005
Cheng JB, Levine M, Bell NH et al (2004) Genetic evidence that the human CYP2R1 enzyme is a key vitamin D 25-hydroxylase. Proc Natl Acad Sci USA 101:7711–7715. https://doi.org/10.1073/pnas.0402490101
Hayashi S, Noshiro M, Okuda K (1986) Isolation of a cytochrome P-450 that catalyzes the 25-hydroxylation of vitamin D3 from rat liver microsomes. J Biochem 99:1753–1763
Gupta RP, He YA, Patrick KS et al (2005) CYP3A4 is a vitamin D-24- and 25-hydroxylase: analysis of structure function by site-directed mutagenesis. J Clin Endocrinol Metab 90:1210–1219. https://doi.org/10.1210/jc.2004-0966
Strushkevich N, Usanov S, Plotnikov AN et al (2008) Structural analysis of CYP2R1 in complex with vitamin D3. J Mol Biol 380:95–106. https://doi.org/10.1016/j.jmb.2008.03.065
Hannemann F, Bichet A, Ewen KM, Bernhardt R (2007) Cytochrome P450 systems—biological variations of electron transport chains. Biochim Biophys Acta 1770:330–344. https://doi.org/10.1016/j.bbagen.2006.07.017
McLean KJ, Sabri M, Marshall KR et al (2005) Biodiversity of cytochrome P450 redox systems. Biochem Soc Trans 33:796. https://doi.org/10.1042/BST0330796
Bisswanger H (2014) Enzyme assays. Perspect Sci 1:41–55. https://doi.org/10.1016/j.pisc.2014.02.005
Voet D, Voet JG (2004) Biochemistry, 3rd edn. Wiley, Hoboken
Chaudhuri R, Cheng Y, Middaugh CR, Volkin DB (2014) High-throughput biophysical analysis of protein therapeutics to examine interrelationships between aggregate formation and conformational stability. AAPS J 16:48–64. https://doi.org/10.1208/s12248-013-9539-6
Acknowledgements
This work was financially supported by Research University Grant (05-02-12-2185RU) from Universiti Putra Malaysia. We acknowledge Malaysia Genome Institute (MGI) for providing the bacterial strain and its genome data. Swi See Ang would like to thank Ministry of Education, Malaysia for MyPhD scholarship award.
Author information
Authors and Affiliations
Contributions
SSA designed, performed the experiments, analyzed the data and prepared the manuscript. ALTC, BAT and MBAR involved in data analysis. YMN and MAF participated in research design, data interpretation and drafted the manuscript. ABS supervised the study, designed the experiment and revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest to disclose regarding the publication of this paper.
Rights and permissions
About this article
Cite this article
Ang, S.S., Salleh, A.B., Chor, L.T. et al. Biochemical Characterization of the Cytochrome P450 CYP107CB2 from Bacillus lehensis G1. Protein J 37, 180–193 (2018). https://doi.org/10.1007/s10930-018-9764-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-018-9764-z