Abstract
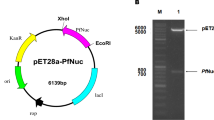
Pfu DNA polymerase (Pfu) is a DNA polymerase isolated from the hyperthermophilic archaeon Pyrococcus furiosus. With its excellent thermostability and high fidelity, Pfu is well known as one of the enzymes widely used in the polymerase chain reaction. In this study, the recombinant plasmid pLysS His6-tagged Pfu-pET28a was constructed. His-tagged Pfu was expressed in Escherichia coli BL21 (DE3) competent cells and then successfully purified with the ÄKTAprime plus compact one-step purification system by Ni2+ chelating affinity chromatography after optimization of the purification conditions. The authenticity of the purified Pfu was further confirmed by peptide mass fingerprinting. A bio-assay indicated that its activity in the polymerase chain reaction was equivalent to that of commercial Pfu and its isoelectric point was found to be between 6.85 and 7.35. These results will be useful for further studies on Pfu and its wide application in the future.







Similar content being viewed by others
Abbreviations
- AP:
-
Ammonium persulfate
- bp:
-
Base pair
- E. coli :
-
Escherichia coli
- EDTA:
-
Ethylene diamine tetraacetic acid
- IPTG:
-
Isopropyl β-d-1-thiogalactopyranoside
- PAGE:
-
Polyacrylamide gel electrophoresis
- pEGFP:
-
Plasmid enhanced green fluorescent protein
- pI:
-
Isoelectric point
- PMSF:
-
Phenylmethylsulfonyl fluoride
- SDS:
-
Sodium dodecyl sulfate
- TEMED:
-
N,N,N′,N′-Tetramethylethylenediamine
- U:
-
Unit
References
Lundberg KS, Shoemaker DD, Adams MWW, Short JM, Sorge JA, Mathur EJ (1991) High-fidelity amplification using a thermostable DNA polymerase isolated from Pyrococcus furiosus. Gene 108:1–6
Ppyun H, Kim I, Cho SS, Seo KJ, Yoon K, Kwon S (2012) Improved PCR performance using mutant Tpa-S DNA polymerases from the hyperthermophilic archaeon Thermococcus pacificus. J Biotechnol 164:363–370
Cline J, Braman JC, Hogrefe HH (1996) PCR fidelity of Pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res 24:3546–3551
Pavlov AR, Pavlova NV, Kozyavkin SA, Slesarev AI (2004) Recent developments in the optimization of thermostable DNA polymerases for efficient applications. Trends in Biotechnol 22:253–260
Kim YJ, Cha S, Lee HS, Ryu YG, Bae SS, Cho Y, Cho H, Kim S, Kwon S, Lee J, Kang SG (2008) Sensing domain and extension rate of a family B-Type DNA Polymerase determine the stalling at a deaminated base. J Microbiol Biotechnol 18:1377–1385
Norholm MHH (2010) A mutant Pfu DNA polymerase designed for advanced uracil-excision DNA engineering. BMC Biotechnol 10:21–27
Jozwiakowski SK, Connolly BA (2009) Plasmid-based lacZa assay for DNA polymerase fidelity: application to archaeal family-B DNA polymerase. Nucleic Acids Res 37:102–108
Melissis S, Labrou NE, Clonis YD (2007) One-step purification of Taq DNA polymerase using nucleotide-mimetic affinity chromatography. J Biotechnol 2:121–132
Lu C, Erickson HP (1997) Expression in Escherichia coli of the thermostable DNA polymerase from Pyrococcus furiosus. Protein Expr Purif 11:179–184
Hu J, Wang F, Liu C (2015) Development of an efficient process intensification strategy for enhancing Pfu DNA polymerase production in recombinant Escherichia coli. Bioprocess Biosyst Eng 38:651–659
Ferralli P, Egan J (2007) Making Taq DNA polymerase in the undergraduate biology laboratory. Bios 78:69–74
Sun ZH, Cai J (2006) Purification of recombinant Pfu DNA polymerase using a new JK110 resin. Korean J Chem Eng 23:607–609
Nika H, Angeletti RH, Hawke DH (2014) N-terminal protein characterization by mass spectrometry using combined microscale liquid and solid-phase derivatization. J Biomol Tech 25:77–86
Uemori T, Ishino Y, Toh H, Asada K, Kato I (1993) Organization and nucleotide sequence of the DNA polymerase gene from the archaeon Pyrococcus furiosus. Nucl Acids Res 21(2):259–265
Ignatov KB, Barsova EV, Fradkov AF, Blagodatskikh KA, Kramarova TV, Kramarov VM (2014) A strong strand displacement activity of thermostable DNA polymerase markedly improves the results of DNA amplification. Biotechniques 57:81–87
Braithwaite DK, Ito J (1993) Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res 21(4):787–802
Kim SW, Kim DU, Kim JK, Kang LW, Cho HS (2008) Crystal structure of Pfu, the high fidelity DNA polymerase from Pyrococcus furiosus. Int J Biol Macromol 42:356–361
Drum M, Kranaster R, Ewald C, Blasczyk R, Marx A (2014) Variants of a Thermus aquaticus DNA polymerase with increased selectivity for applications in allele- and methylation-specific amplification. PLoS ONE 9(5):e96640–e96650
Biles BD, Connolly BA (2004) Low-fidelity Pyrococcus furiosus DNA polymerase mutants useful in error-prone PCR. Nucleic Acids Res 32(22):176–182
Takagi M, Nishioka M, Kakihara H, Kitabayashi M, Inoue H, Kawakami B, Oka M, Imanaka T (1997) Characterization of DNA polymerase from Pyrococcus sp. strain KOD1 and its application to PCR. Appl Environ Microbiol 63(11):4504–4510
Kim YJ, Lee HS, Bae SS, Jeon JH, Lim JK, Cho Y, Nam KH, Kang SG, Kim SJ, Kwon ST, Lee JH (2007) Cloning, purification, and characterization of a new DNA polymerase from a hyperthermophilic archaeon, Thermococcus sp. NA1. J Microbiol Biotechnol 17(7):1090–1097
Mroczkowski BS, Huvar A, Lernhardt W, Misono K, Nielson K, Scott B (1994) Secretion of thermostable DNA polymerase using a novel baculovirus vector. J Biol Chem 269(18):13522–13528
Miura M, Tanigawa C, Fujii Y, Kaneko S (2013) Comparison of six commercially-available DNA polymerase for direct PCR. Rev Inst Med Trop Sao Paulo 55(6):401–406
Viennois E, Chen F, Laroui H, Baker MT, Merlin D (2013) Dextran sodium sulfate inhibits the activities of both polymerase and reverse transcriptase: lithium chloride purification, a rapid and efficient technique to purify RNA. BMC Res Notes 6:360–367
Chong SS, Eichler EE, Nelson DL, Hughes MR (1994) Robust amplification and ethidium-visible detection of the fragile X syndrome CGG repeat using Pfu polymerase. Am J Med Genet 51(4):522–526
Eckert KA, Kunkel TA (1991) DNA polymerase fidelity and the polymerase chain reaction. Genome Res 1:17–24
Moser IJ, DiFrancesco RA, Gowda K, Klingele AJ, Sugar DR, Stocki S, Mead DA, Schoenfeld TW (2012) Thermostable DNA polymerase from a viral metagenome is a potent RT-PCR enzyme. PLoS ONE 7(6):e38371–e38383
Barnes DE, Lindahl T (2004) Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet 38:445–476
Krokan HE, Drablos F, Slupphaug G (2002) Uracil in DNA—occurrence, consequences and repair. Oncogene 21(58):8935–8948
Melissis S, Labrou NE, Clonis YD (2006) Nucleotide-mimetic synthetic ligands for DNA-recognizing enzymes one-step purification of Pfu DNA polymerase. J Chromatogr A 1122(1–2):63–75
Pearl LH (2000) Structure and function in the uracil-DNA glycosylase superfamily. Mutat Res/DNA Repair 460(3–4):165–181
Wardle J, Burgers PMJ, Cann IKO, Darley K, Heslop P, Johansson E, Lin LJ, McGlynn P, Sanvoisin J, Stith CM, Connolly BA (2008) Uracil recognition by replicative DNA polymerases is limited to the archaea, not occurring with bacteria and eukarya. Nucleic Acids Res 36(3):705–711
Dabrowski S, Ahring BK (2000) Cloning, expression, and purification of the His6-tagged hyper-thermostable dUTPase from Pyrococcus woesei in Escherichia coli: application in PCR. Protein Expr Purif 19(1):107–112
Niimi H, Mori M, Tabata H, Minami H, Ueno T, Hayashi S, Kitajima I (2011) A novel eukaryote-made thermostable DNA polymerase which is free from bacterial DNA contamination. J Clin Microbiol 49(9):3316–3320
Kuroita T, Matsumura H, Yokota N, Kitabayashi M, Hashimoto H, Inoue T, Imanaka T, Kai Y (2005) Structural mechanism for coordination of proofreading and polymerase activities in archaeal DNA polymerases. J Mol Biol 351(2):291–298
Spangler R, Goddard NL, Thaler DS (2009) Optimizing Taq polymerase concentration for improved signal-to-noise in the broad range detection of low abundance bacteria. PLoS ONE 4(9):e7010–e7017
Acknowledgments
We would like to thank the Natural Basic Sciences Personnel Training Foundation of China (No. J1310033) for funding support. We acknowledged Professor Jijie Chai (National Institute of Biological Sciences of Beijing) for the provision of Pfu gene.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Rights and permissions
About this article
Cite this article
Zheng, W., Wang, Q. & Bi, Q. Construction, Expression, and Characterization of Recombinant Pfu DNA Polymerase in Escherichia coli . Protein J 35, 145–153 (2016). https://doi.org/10.1007/s10930-016-9651-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-016-9651-4