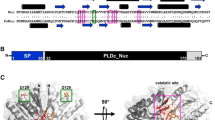
Abstract
Non-specific endonucleases can be used for the digestion of nucleic acids because they hydrolyze DNA/RNA into 3–5 base pairs (bp) length oligonucleotide fragments without strict selectivity. In this work, a novel non-specific endonuclease from Pseudomonas fluorescens (PfNuc) with high activities for both DNA and RNA was successfully cloned and expressed in Escherichia coli. The production of PfNuc in flask scale could be achieved to 1.73 × 106 U/L and 4.82 × 106 U/L for DNA and RNA by investigation of the culture and induction conditions. The characterization of PfNuc indicated that it was Mg2+-dependent and the catalytic activity was enhanced by 3.74 folds for DNA and 1.06 folds for RNA in the presence of 5 mM Mg2+. The specific activity of PfNuc for DNA was 1.44 × 105 U/mg at pH 8.0 and 40 °C, and 3.93 × 105 U/mg for RNA at pH 8.5 and 45 °C. The Km of the enzyme for both DNA and RNA was close to 43 µM. The Vmax was 6.40 × 105 U/mg and 1.11 × 106 U/mg for DNA and RNA, respectively. There was no observed activity loss when PfNuc was stored at 4 °C and − 20 °C after 28 days or 10 repeated freeze–thaw cycles at − 80 °C. Molecular docking revealed that PfNuc formed 17 and 19 hydrogen bonds with single-stranded RNA and double-stranded DNA, respectively. These results could explain the high activity and stability of PfNuc, suggesting its great potential applications in the industry and clinic.








Similar content being viewed by others
Data availability
No datasets were generated or analyzed during the current study.
References
Asano Y, Matsumoto Y, La Rose J et al (2021) Endonuclease increases efficiency of osteoblast isolation from murine calvariae. Sci Rep 11:8502. https://doi.org/10.1038/s41598-021-87716-8
Benedik MJ, Strych U (1998) Serratia marcescens and its extracellular nuclease. FEMS Microbiol Lett 165:1–13. https://doi.org/10.1016/S0378-1097(98)00234-1
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 132:150–156
Cao K, Zhang Y, Yao Q et al (2022) Hypericin blocks the function of HSV-1 alkaline nuclease and suppresses viral replication. J Ethnopharmacol 296:115524. https://doi.org/10.1016/j.jep.2022.115524
Chen LY, Ho HC, Tsai YC, Liao TH (1993) Deoxyribonuclease of Syncephalastrum racemosum-enzymatic properties and molecular structure. Arch Biochem Biophys 303:51–56. https://doi.org/10.1006/abbi.1993.1254
Chen C, Krause K, Pettitt BM (2009) Advantage of being a dimer for Serratia marcescens endonuclease. J Phys Chem B 113:511–521. https://doi.org/10.1021/jp8057838
Cote J, Ruiz-Carrillo A (1993) Primers for mitochondrial DNA replication generated by endonuclease G. Science 261:765–769. https://doi.org/10.1126/science.7688144
Filimonova M, Gubskaya V, Nuretdinov I et al (2001) Study of the mechanism of action of p-chloromercuribenzoate on endonuclease from the bacterium Serratia marcescens. Biochem Mosc 66:323–327. https://doi.org/10.1023/A:1010212232287
Friedhoff P, Gimadutdinow O, Pingoud A (1994) Identification of catalytically relevant amino acids of the extracellular Serratia marcescens endonuclease by alignment-guided mutagenesis. Nucleic Acids Res 22:3280–3287. https://doi.org/10.1093/nar/22.16.3280
Friedhoff P, Kolmes B, Gimadutdinow O et al (1996) Analysis of the mechanism of the Serratia nuclease using site-directed mutagenesis. Nucleic Acids Res 24:2632–2639. https://doi.org/10.1093/nar/24.14.2632
Fujimoto M, Kuninaka A, Yoshino H (1974) Purification of a nuclease from Penicillium citrinum. Agric Biol Chem 38:777–783. https://doi.org/10.1080/00021369.1974.10861230
Green AP, Huang JJ, Scott MO et al (2002) A new scalable method for the purification of recombinant adenovirus vectors. Hum Gene Ther 13:1921–1934. https://doi.org/10.1089/10430340260355338
Guo L-B, Zhu C-Y, Wu Y-B et al (2021) A novel chondroitin AC lyase from Pedobacter xixiisoli: Cloning, expression, characterization and the application in the preparation of oligosaccharides. Enzyme Microb Technol 146:109765. https://doi.org/10.1016/j.enzmictec.2021.109765
Li L, Rohrmann GF (2000) Characterization of a baculovirus alkaline nuclease. J Virol 74:6401–6407. https://doi.org/10.1128/JVI.74.14.6401-6407.2000
Li S-M, Bai F-L, Xu W-J et al (2014) Removing residual DNA from Vero-cell culture-derived human rabies vaccine by using nuclease. Biologicals 42:271–276. https://doi.org/10.1016/j.biologicals.2014.06.005
Li F, Shi Y, Zhang J et al (2018) Cloning, expression, characterization and homology modeling of a novel water-forming NADH oxidase from Streptococcus mutans ATCC 25175. Int J Biol Macromol 113:1073–1079. https://doi.org/10.1016/j.ijbiomac.2018.03.016
Liu J, Li Z, Li J, Liu Z (2019) Application of Benzonase in preparation of decellularized lamellar porcine corneal stroma for lamellar keratoplasty. J Biomed Mater Res 107:2547–2555. https://doi.org/10.1002/jbm.a.36760
Liu C, Su W, Guo L, Zhang Y (2020) Cloning, expression, and characterization of a novel heparinase I from Bacteroides eggerthii. Prep Biochem Biotechnol 50:477–485. https://doi.org/10.1080/10826068.2019.1709977
MacLellan SR, Forsberg CW (2001) Properties of the major non-specific endonuclease from the strict anaerobe Fibrobacter succinogenes and evidence for disulfide bond formation in vivo. Microbiology 147:315–323
Meiss G, Franke I, Gimadutdinow O et al (1998) Biochemical characterization of Anabaena sp. strain PCC 7120 non-specific nuclease NucA and its inhibitor NuiA. Eur J Biochem 251:924–934. https://doi.org/10.1046/j.1432-1327.1998.2510924.x
Morris GM, Huey R, Lindstrom W et al (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. https://doi.org/10.1002/jcc.21256
Muro-Pastor AM, Flores E, Herrero A, Wolk CP (1992) Identification, genetic analysis and characterization of a sugar-non-specific nuclease from the cyanobacterium Anabaena sp. PCC 7120. Mol Microbiol 6:3021–3030. https://doi.org/10.1111/j.1365-2958.1992.tb01760.x
Nierzwicki Ł, East KW, Binz JM et al (2022) Principles of target DNA cleavage and the role of Mg2+ in the catalysis of CRISPR–Cas9. Nat Catal 5:912–922. https://doi.org/10.1038/s41929-022-00848-6
Pei J, Wei W, Wang D et al (2023) Cloning, expression, and characterization of a highly stable heparinase I from Bacteroides xylanisolvens. Polymers 15:1776. https://doi.org/10.3390/polym15071776
Pingoud A, Fuxreiter M, Pingoud V, Wende W (2005) Type II restriction endonucleases: structure and mechanism. Cell Mol Life Sci 62:685–707. https://doi.org/10.1007/s00018-004-4513-1
Pingoud V, Wende W, Friedhoff P et al (2009) On the divalent metal ion dependence of DNA cleavage by restriction endonucleases of the EcoRI Family. J Mol Biol 393:140–160. https://doi.org/10.1016/j.jmb.2009.08.011
Pontius J, Richelle J, Wodak SJ (1996) Deviations from standard atomic volumes as a quality measure for protein crystal structures. J Mol Biol 264:121–136. https://doi.org/10.1006/jmbi.1996.0628
Puyet A, Greenberg B, Lacks SA (1990) Genetic and structural characterization of endA: A membrane-bound nuclease required for transformation of Streptococcus pneumoniae. J Mol Biol 213:727–738. https://doi.org/10.1016/S0022-2836(05)80259-1
Robert X, Gouet P (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. https://doi.org/10.1093/nar/gku316
Schmitz S, Nölle V, Elleuche S (2019) A non-specific nucleolytic enzyme and its application potential in EDTA-containing buffer solutions. Biotechnol Lett 41:129–136. https://doi.org/10.1007/s10529-018-2618-0
Schmitz S, Wieczorek M, Nölle V, Elleuche S (2020) Characterization of single amino acid variations in an EDTA-tolerating non-specific nuclease from the ice-nucleating bacterium Pseudomonas syringae. Mol Biotechnol 62:67–78. https://doi.org/10.1007/s12033-019-00229-8
Schwardmann LS, Schmitz S, Nölle V, Elleuche S (2019) Decoding essential amino acid residues in the substrate groove of a non-specific nuclease from Pseudomonas syringae. Catalysts 9:941. https://doi.org/10.3390/catal9110941
Song Q, Zhang X (2008) Characterization of a novel non-specific nuclease from thermophilic bacteriophage GBSV1. BMC Biotechnol 8:43. https://doi.org/10.1186/1472-6750-8-43
Vafina G, Zainutdinova E, Bulatov E, Filimonova MN (2018) Endonuclease from gram-negative bacteria Serratia marcescens is as effective as Pulmozyme in the hydrolysis of DNA in sputum. Front Pharmacol 9:114. https://doi.org/10.3389/fphar.2018.00114
Vincent RD, Hofmann TJ, Zassenhaus HP (1988) Sequence and expression of NUC1, the gene encoding the mitochondrial nuclease in Saccharomyces cerevisiae. Nucleic Acids Res 16:3297–3312. https://doi.org/10.1093/nar/16.8.3297
Waterhouse A, Bertoni M, Bienert S et al (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303. https://doi.org/10.1093/nar/gky427
Wu C-C, Lin JLJ, Yuan HS (2020) Structures, mechanisms, and functions of His-Me finger nucleases. Trends Biochem Sci 45:935–946. https://doi.org/10.1016/j.tibs.2020.07.002
Yang W (2011) Nucleases: diversity of structure, function and mechanism. Quarterl Rev Biophys 44:1–93. https://doi.org/10.1017/S0033583510000181
Yonemura K, Matsumoto K, Maeda H (1983) Isolation and characterization of nucleases from a clinical isolate of Serratia marcescens kums 3958 1. J Biochem 93:1287–1295. https://doi.org/10.1093/oxfordjournals.jbchem.a134262
Yuan J, Amend A, Borkowski J et al (1999) MULTICLUSTAL: a systematic method for surveying Clustal W alignment parameters. Bioinformatics 15:862–873. https://doi.org/10.1093/bioinformatics/15.10.862
Acknowledgements
We appreciated the financial support from the Guangxi Natural Science Foundation (2021GXNSFAA196005) and the National Natural Science Foundation of China (No. 22068001).
Funding
Natural Science Foundation of Guangxi Province, 2021GXNSFAA196005, National Natural Science Foundation of China, 22068001.
Author information
Authors and Affiliations
Contributions
Ke-Ke Han: Investigation, Writing—Original draft preparation; Qiang Zhou: Methodology, Investigation, Writing—Original draft preparation; Yang-Nan Li: Investigation; Miao Tian: Investigation; Jing-Yi Zhang: Investigation; Ye-Wang Zhang: Conceptualization, Writing— Reviewing and Editing, Supervision.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by Yusuf Akhter.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Han, KK., Zhou, Q., Tian, M. et al. Cloning, heterologous expression, and molecular characterization of a highly active and stable non-specific endonuclease from Pseudomonas fluorescens. Arch Microbiol 206, 125 (2024). https://doi.org/10.1007/s00203-024-03867-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-024-03867-y