Abstract
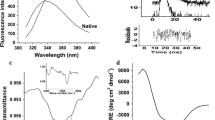
The kinetically stable alkaline serine protease from Nocardiopsis sp.; NprotI, possessing polyproline II fold (PPII) was characterized for its pH stability using proteolytic assay, fluorescence and Circular Dichroism (CD) spectroscopy, and Differential Scanning Calorimetry (DSC). NprotI was found to be functionally stable when incubated at pH 1.0, even after 24 h, while after incubation at pH 10.0, drastic loss in the activity was observed. The enzyme showed enhanced activity after incubation at pH 1.0 and 3.0, at higher temperature (50–60 °C). NprotI maintained the overall PPII fold in broad pH range as seen using far UV CD spectroscopy. The PPII fold of NprotI incubated at pH 1.0 remained fairly intact up to 70 °C. Based on the isodichroic point and Tm values revealed by secondary structural transitions, different modes of thermal denaturation at pH 1.0, 5.0 and 10.0 were observed. DSC studies of NprotI incubated at acidic pH (pH 1.0–5.0) showed Tm values in the range of 74–76 °C while significant decrease in Tm (63.8 °C) was observed at pH 10.0. NprotI could be chemically denatured at pH 5.0 (stability pH) only with guanidine thiocynate. NprotI can be classified as type III protein among the three acid denatured states. Acid tolerant and thermostable NprotI can serve as a potential candidate for biotechnological applications.




Similar content being viewed by others
Abbreviations
- PPII:
-
Polyproline II
- CD:
-
Circular Dichroism
- DSC:
-
Differential Scanning Calorimetry
- NprotI:
-
Nocardiopsis protease I
- GuSCN:
-
Guanidine thiocyanate
- GdnHCl:
-
Guanidine hydrochloride
- MRE:
-
Mean residual ellipticity
- ANS:
-
1-Anilino 8-napthalene sulfonic acid
- Napase:
-
Nocardiopsis alba protease
References
Cunningham EL, Jaswal SS, Sohl JL, Agard DA (1999) Kinetic stability as a mechanism for protease longevity. Proc Natl Acad Sci 96:11008–11014
Yadav SC, Jagannadham MV (2009) Complete conformational stability of kinetically stable dimeric serine protease milin against pH, temperature, urea, and proteolysis. Eur Biophys J 38:981–990
Rohamare SB, Dixit VS, Nareddy PK, Sivaramakrishna D, Swamy MJ, Gaikwad SM (2013) Polyproline fold—in imparting kinetic stability to an alkaline serine endopeptidase. Biochim Biophys Acta 1834:708–716
Kelch BA, Eagen KP, Erciyas FP, Humphris EL, Thomason AR, Mitsuiki S, Agard DA (2007) Structural and mechanistic exploration of acid resistance: kinetic stability facilitates evolution of extremophilic behavior. J Mol Biol 368:870–883
Matthew JB (1985) Electrostatic effects in proteins. Annu Rev Biophys Chem 14:387–417
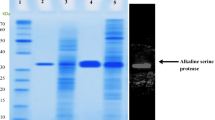
Dixit VS, Pant A (2000) Comparative characterization of two serine endopeptidases from Nocardiopsis sp. NCIM 5124. Biochim Biophys Acta 1523:261–268
Qi PX, Wickham ED, Farrell HM Jr (2004) Thermal and alkaline denaturation of bovine β-casein. Protein J 23:389–402
Rucker AL, Creamer TP (2002) Polyproline II helical structure in protein unfolded states: lysine peptides revisited. Protein Sci 11:980–985
Shimizu A, Kawai K, Yanagino M, Wakiyama T, Machida M, Kameyama K, Naito Z (2007) Characteristics of type IV collagen unfolding under various pH conditions as a model of pathological disorder in tissue. J Biochem 142:33–40
Ptitsyn OB (1987) Protein folding: hypotheses and experiments. J Protein Chem 6:277–293
Ewbank JJ, Creighton TE (1991) The molten globule protein conformation probed by disulphide bonds. Nature 350:518–520
Buchner J, Renner M, Lilie H, Hinz HJ, Jaenicke R, Kiefhaber T, Rudolph R (1991) Alternatively folded states of an immunoglobulin. Biochemistry 30:6922–6929
Haq SK, Ahmad MF, Khan RH (2003) The acid-induced state of glucose oxidase exists as a compact folded intermediate. Biochem Biophys Res Commun 303:685–692
Conejero-Lara F, Azuaga AI, Mateo PL (1997) Differential scanning calorimetry of thermolysin and its 255–316 and 205–316 C-terminal fragments. React Funct Polym 34:113–120
Zafrullah M, Khursheed Z, Yadav S, Sahgal D, Jameel S, Ahmad F (2004) Acidic pH enhances structure and structural stability of the capsid protein of hepatitis E virus. Biochem Biophys Res Commun 313:67–73
Fink AL, Calciano LJ, Goto Y, Kurotsu T, Palleros DR (1994) Classification of acid denaturation of proteins: intermediates and unfolded states. Biochemistry 33:12504–12511
Rohamare SB, Gaikwad SM (2014) Tryptophan environment and functional characterization of a kinetically stable serine protease containing a polyproline II fold. J Fluoresc 24:1363–1370
Acknowledgments
The authors thank Dr. M. Fernandes, Organic Chemistry Division, NCL, for allowing the use of CD spectropolarimeter. SR, SD and PKN were supported by Senior Research Fellowships from the CSIR, New Delhi, India. The MicroCal VP-DSC equipment used in this study was supported by a UPE grant from the UGC (India) to the University of Hyderabad.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10930_2014_9597_MOESM3_ESM.doc
Figure S3: Activity profile of NprotI incubated with GuSCN (0-6 M). Experiment was performed as described in Materials and Methods section (DOC 67 kb)
Rights and permissions
About this article
Cite this article
Rohamare, S., Javdekar, V., Dalal, S. et al. Acid Stability of the Kinetically Stable Alkaline Serine Protease Possessing Polyproline II Fold. Protein J 34, 60–67 (2015). https://doi.org/10.1007/s10930-014-9597-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-014-9597-3