Abstract
Eighty-seven bacteria isolates were screened for the presence of the levansucrase encoding gene and those possessing the gene were used in gene expression studies and the production of levan. Based on both screening methods, isolate P19-1 was determined as the producer bacterium with the highest amounts of levan. Whole genome sequencing was done to characterize the isolate and determine its genomic potential. Both its 16 S rRNA and genome were closest to Chromohalobacter salexigens. The levansucrase gene and genes suggesting a versatile carbohydrate and nitrogen metabolism were annotated. A secondary metabolism suggestive of a possible new role in promoting plant growth in hypersaline environments was revealed. The chemical structure analysis of the produced polymer was confirmed by comparison with commercial pure Erwinia herbicola levan. Also, C. salexigens levan showed high biocompatibility. Overall, this is a practical molecular approach that will help researchers to effectively screen microbes with levan production potential.
Similar content being viewed by others
Introduction
Levan is a type of fructan homopolymer composed of β-fructose units [1]. Lippmann proposed the first accepted definition of this substance in 1881 and used the term “lavulan” to describe the gum obtained from molasses [2]. Subsequently, the term “levan” was introduced by Greig-Smith in 1901 [3]. These definitions paved the way for research on the biosynthesis and collection of levan, leading to the development of the polysaccharide trade [4].
Levan can be synthesized by various organisms, including plants, yeasts, fungi, and bacteria. The levan biosynthesis is generally carried out by the enzyme levansucrase (EC 2.4.1.10), although the formation of levansucrase is more commonly observed in bacteria. The production of microbial levan is influenced by various factors, including temperature, pH, oxygen concentration, and nutrient content [1]. Moreover, levan polymers synthesized by different microorganisms exhibit variations in their molecular weight, viscosity, and bioactivity, which are attributed to the producer microorganism and its growing parameters, such as the amount of carbon and nitrogen sources [1]. To achieve high efficiency in microbial levan production, different microorganisms and optimum production conditions that enhance the levan synthesis of the microorganism are tested [1].
Levan is characterized by its high adhesiveness and low internal viscosity, which are important features that distinguish it from other polymers [5]. Due to its unique physical and chemical properties and its biological activities, levan has garnered significant attention and is being studied for its potential applications in various industrial sectors, including pharmaceuticals, food, and cosmetics [5].
Despite successful microbial levan production in laboratory environments, large-scale production of this product has not yet been fully achieved [6]. Several significant reasons hinder the industrial-scale production of levan. Firstly, levansucrase requires sucrose as a substrate for levan synthesis, and the supply and cost of sucrose on a large scale can be limiting factors. Additionally, it can be challenging to maintain stable conditions such as temperature and pH, which can affect the functioning of the levansucrase enzyme on a large scale. Optimizing these conditions can be time-consuming and costly. Finally, purification, a crucial step in levan production, can be complex and time-consuming on a large scale, adding to the overall cost of production [7]. Consequently, studies on levan production from different microorganisms are ongoing. Levan production has been observed in mesophilic species belonging to genera such as Bacillus, Pseudomonas, and Zymomonas, as well as in halophilic species [8]. Levan production by halophilic isolates is advantageous for large-scale production, as it enables production in non-sterile conditions at high salinity levels [9].
Currently, there are two main approaches for discovering new microbial levan producers. The conventional way involves screening for high producers [10], while the systems-based approach applies metabolic engineering principles from a holistic perspective. Over the years the former approach revolved around growing the candidate isolates on suitable substrates and observing slime mucoid phenotype typical of levan production [11]. The activation of enzymes responsible for the synthesis of exopolysaccharides such as levan is highly dependent on environmental conditions. Therefore, these traditional methods may exclude potential producers that are not grown in optimal conditions [12]. Identifying these strains could provide a more accurate pool of potential producer strains that could be further optimized for culture conditions using experimental design methodologies such as response surface methodology and statistical analysis. In this context, it is desirable to identify genotypic potential rather than phenotype alone.
Levansucrases (EC 2.4.1.10) are the group of enzymes responsible for the synthesis of levan from sucrose as substrate. Levan-producing microbes have been shown to express the levansucrase gene which catalyzes the levan biosynthesis from sucrose, and hydrolysis of levan to fructose units [13]. The presence of this gene in levan producers suggests it could be used as a marker for levan biosynthesis. Dedicated to levan production, levansucrase is often favored for industrial applications. Therefore, microbes expressing this gene are likely to be favored for industrial applications [14].
Quantitative PCR (Q-PCR), also known as real-time PCR, is a widely used technique in microbial ecology for determining the abundance and expression of taxonomic and functional gene markers. This approach can be applied at both environmental and bacterial levels and is typically combined with traditional PCR and fluorescent detection techniques in real time. Q-PCR and Reverse Transcriptase Q-PCR (RT-Q-PCR) have significant applications in both theoretical and practical implementations, as they enable the detection of amplicons during the early exponential phase. These techniques are valuable tools for studying microbial communities and their functions, and they have numerous applications in fields such as environmental monitoring, medical diagnostics, and biotechnology [15].
Relative quantification is a method that expresses the correlative change at the mRNA levels. This approach detects variations in steady-state mRNA levels of a gene in various samples and expresses it with the correlation of the levels of another RNA in these samples. Relative quantification is a useful tool for investigating small physiological changes in gene expression. Fixed expressed reference genes are selected for reference, and they can be co-amplified in the same tube (endogenous controls) or amplified in a separate tube (exogenous controls) [16]. Therefore, this screening approach, namely quantitative PCR, is considered practical in microbial diagnostics and microbial ecology and biotechnological applications related to biopolymer studies. Relative quantification is a valuable technique for studying gene expression and can provide insights into the molecular mechanisms underlying various biological processes.
In this study, the levan production potential of halophilic bacterial species isolated from saline environments was investigated. Initially, levansucrase gene amplification was studied in all isolates. Microorganisms with positive bands were then used for gene expression studies using relative quantitative Real-Time PCR. Simultaneously, levan production abilities were validated by carrying out microbial production. At the outset of the study, it was hypothesized that microorganisms with higher gene expression would produce higher amounts of levan. At the end of the study, the levan production of the microorganism with the highest production potential was optimized in a fermenter, and the resulting biopolymer was characterized. In addition, whole genome analysis of the selected microorganism was performed. This study provides valuable insights into the levan production potential of halophilic bacterial species and highlights the importance of gene expression studies in predicting levan production capabilities.
Materials and Methods
Bacterial Strains
The halophilic bacterial strains used in this study were isolated from hypersaline areas (Tuz Lake, Çamaltı Saltern, Tunceli Pülümür Spring Saltern) in different geographical regions of Turkey. Isolated halophilic strains were grown in a modified growth medium (5,6 g/L MgCl2; 14 g/L MgSO4; 2,8 g/L KCl; 0,32 g/L NaBr; 0,08 g/L NaHCO3; 2 g/L CaCl2; 1 g/L yeast extract; 1 g/L peptone) containing 12% and 18% salt at 37 °C for 24 h. The long-term storage was carried out in glycerol at -80 °C.
Amplification of the Gene Region Encoding the Levansucrase Enzyme
The gene region encoding the levansucrase enzyme was referenced by Halomonas smyrnensis [17]. The DNAs of the halophilic isolates were extracted following the A.B.T™ DNA Purification Kit procedure and a polymerase chain reaction was set up for levansucrase gene region amplification consisting of pre-denaturation at 95 ℃ for 5 min then 35 cycles at 95 ℃ for 30 s, 60 ℃ for 30 s and 72 ℃ for 75 s. Primers used in the amplification of the gene coding for levansucrase (sacB) designed specifically for the reference gene region were used in the reaction (Forward:5′-CGG TGA TGT GGT ATC AGT CG-3′; Reverse:5′-CTT GCC ACC TTT CTC TAC GC-3′). The products formed at the end of the PCR reaction were visualized by running gel electrophoresis. Positive results showing bands with the size of the levansucrase gene region were sequenced. Multiple alignment of the sequences obtained was done in MEGA11 (11.0.10) software and isolates with conserved regions associated with the levansucrase gene region were selected.
RNA Isolation and cDNA Synthesis for Expression Analysis of the Gene Encoding Levansucrase
Real-time PCR reactions were carried out using cDNAs as templates. In order to determine the expression levels of the isolates associated with the levansucrase gene region, RNA isolation was first performed. The A.B.T™ Blood/Tissue RNA Purification kit procedure was followed for RNA isolation and A.B.T™ High-Capacity cDNA Reverse Transcription for cDNA synthesis.
Relative Quantification Real-Time PCR for Levansucrase
PCR Relative quantification method is based on the signal which is obtained from the target. The 2−ΔΔCT method is convenient to analyze the expression level of the target gene. Designed primer pairs and universal primer pairs for 16 S rRNA were used for the relative quantitative 2−ΔΔCT method [18]. All reactions were performed in Roche, LightCycler 96. The total volume of the PCR reaction mix was 20 µL made up of SYBR Green master mix 1x, forward and reverse primer 10 µM, and 2 µL template cDNA. For each assay, all samples were run in triplicate. The thermal cycling conditions were 40 cycles of 95 °C for 15 s 60 °C for 30 s and 72 °C for 30 s. The relative expression levels were calculated using the 2−ΔΔCT method of Livak and Schmittgen [18].
Levan Production for Validation
After expressing the gene coding levansucrase, levan production was carried out with bacterial isolates for validation. For this purpose, isolates were inoculated in basal medium (pH 7.0) (per liter: 137.2 g NaCl; 50 g sucrose; 7 g K2HPO4; 2 g KH2PO4; 1 g (NH4)2SO4; 0.1 g MgSO4.7H2O, 0.32 g; 0.5 g peptone) [19] then incubated at 37 °C with shaking at 180 rpm for 72 h. This medium was also used for fermenter studies.
Whole-Genome Sequencing
The genomic DNA of P19-1 bacterial strain was extracted, and the purity and integrity were checked as previously described [20]. For DNA sample preparation, 1.0 µg of total DNA was used as the starting material. Sequence libraries were created using the NEBNext® DNA Library Prep Kit, following the indices added to each sample as instructed by the manufacturer. For genomic DNA sequencing, DNA fragments were sheared to a random 350 bp size, then the ends of the DNA fragments were blunted, A-tailed, and ligated to the NEBNext adapter, followed by enrichment PCR with P5 and indexed P7 oligos. The PCR products were purified (AMPure XP system) and the libraries were analyzed for size distribution using the Agilent 2100 Bioanalyzer and quantified by real-time PCR. The genomic DNA was sequenced using the Illumina HiSeq4000 platform.
Genome Assembly, Quality Assessment, Gene Annotation, and Functional Analysis
The obtained sequencing reads were subjected to quality control using FASTQc v0.12.0 [21]. Adapters and low-quality reads were trimmed using Trimmomatic v0.39 [22]. The quality-filtered reads were submitted to the Bacterial and Viral Bioinformatics Resource Center (BV-BRC) v3.30.5i service [23] for genome assembly, annotation, exploration, and analysis. Reads were assembled using Unicycler v0.4.8 [24]. The resulting contigs were evaluated for quality and completeness using QUality ASsessment Tool (QUAST) v5.2.0 [25]. The genome was annotated with the RAST tool kit v3.6.9 (RASTtk) [26].
The predicted open reading frames (ORFs) were also functionally annotated using PROKKA (PROKaryotic Genome Annotation) [27]. The presence of conserved protein domains and functional classifications of the predicted genes were determined using eggNOG-mapper v2 (evolutionary genealogy of genes: Non-supervised Orthologous Groups) database [28].
Secondary Metabolite Gene Cluster Analysis
The identification and characterization of secondary metabolite gene clusters were performed using antibiotics and Secondary Metabolite Analysis Shell (antiSMASH) v7.0 [29]. The software was employed to predict the biosynthetic gene clusters involved in the production of secondary metabolites, including antibiotics and other bioactive compounds.
Pathway and Functional Enrichment Analysis
To elucidate the metabolic pathways present in the bacterial genome, the annotated genes were mapped to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database [30]. The KAAS (KEGG Automatic Annotation Server) tool [31] was utilized for the automatic annotation of the genes to KEGG orthologs.
Comparative Genomics
To assess the genomic relatedness and identify similar sequences, a comparison of the levansucrase gene and the 16 S rRNA from the assembled genome against publicly available bacterial genomes was performed using Basic Local Alignment Search Tool nucleotide (BLASTn) (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The obtained results were further analyzed to identify conserved genomic regions and potential homologous genes.
Data Analysis and Visualization
The obtained results were analyzed using various bioinformatics tools and custom scripts. Data visualization was performed using Proksee (https://proksee.ca) and R programming to generate figures and diagrams representing the genomic features and functional annotations.
Purification and Quantification of Levan
Levan was purified using several procedures, as described by Erdal Altıntas et al. [32]. In the determination of the levan amount, levan was hydrolyzed completely to fructose described by Viikari [33]. Then, the fructose concentration was determined by the dinitrosalicylic acid (DNS) method [34].
Fermentation Experiments
Microorganism and Culture Conditions
Effective strain in producing higher levan was used in fermentation experiments. The aerobic fermentations were carried out batch-wise in 2 L Pyrex flasks containing 1.5 L of working volume, placed on orbital shakers (Infors HT-Minifors 2) under an aeration rate of 1.0 vvm during an incubation time. The inoculum utilized was 5% (v/v) of the total fermented volume.
Fermentation Follow-Up
At the end of the fermentation process, samples separated from the biomass were subjected to ethanol precipitation and dialysis to perform experimental sets. After each batch production was carried out under the conditions specified in the design experimental program. The samples taken from the fermentation medium were centrifuged at 10,000 rpm for 15 min and the biomass was separated from the medium. The separated biomass was dried at 50 °C, and weighed, and the amount of biomass in the 50 mL fermentation medium was determined [19].
The basis for the determination of levansucrase activity; is determining the amount of reducing sugar released as a result of enzymatic hydrolysis by using sugar as a substrate by the DNS method. One unit of levansucrase activity was defined as the amount of enzyme that releases 1 µmol of reducing sugar per minute under standard conditions [35]. Levansucrase enzyme activity was determined using the glucose standard curve with Eq. 1 given below.
A = mean sample absorbance - blank absorbance, c = coefficient from the standard equation, t = incubation time, D = enzyme dilution rate.
To use in levansucrase specific activity calculation, the amount of protein in the fermentation supernatant was measured according to the Bradford [36] method using BSA as the standard. All enzyme and protein assays were done in triplicates.
Experimental Design and Statistical Analysis
The statistical analysis of the data was performed using Design Expert Software (Release 13.00). Optimization of the three most significant variables (carbon source concentration levels 30–60 g/L, agitation rate levels 150–450 rpm, and incubation time levels 24–72 h) was carried out by central composite design (CCD) for levan production. The data of the factors were chosen after a series of preliminary experiments. Twenty experiments were conducted using a face-central composite statistical design (α = 1.5) for the study of three factors each at three levels. The levels were − 1, 0, and + 1 where 0 corresponded to the central point. All experiments were conducted in triplicates and the mean value of levan yield (mg/L) was taken as the response. Quadratic regression analysis of the data was carried out to obtain an empirical model. The optimum levels of all three independent variables were determined by solving the regression equation and analyzing the response surface contour plots. In addition, a 3D Surface was constructed for visual observation of the trend of maximum responses and the interactive effects of the significant variables on the response [19].
Chemical Characterization
Monomeric sugar analysis of acid-hydrolyzed levan was carried out using Thermo Scientific HPLC system, model Ultimate 3000 by estimating the retention time of lyophilized levan (obtained from the best producer) compared with commercial Erwinia herbicola levan from Sigma-Aldrich L8647. Moreover, each of the standard solutions of sugars (fructose, mannose, glucose, and galactose) at experimental conditions using a carbohydrate analysis column (Agilent ZORBAX) with dimensions 4.6 mm and particle size 5 microns in a mobile phase consisting of acetonitrile: distilled water (80:20) (v: v), respectively by injection of 20 µL at a flow rate of 1.4 mL/min and the column temperature is 30 °C [19].
The structural integrity of the obtained levan polymer was investigated by Fourier Transform – Infrared Spectroscopy (Perkin Elmer Spectrum Two) in the 400–4000 wavenumber (cm− 1) range. Commercial levan from Sigma-Aldrich (St. Louis, US) produced by Erwinia herbicola was used as a control [19].
This obtained biopolymer’s binding and molecular properties were determined by proton 1H-NMR spectroscopy at 27 ℃. Commercial levan from Sigma-Aldrich (St. Louis, US) produced by Erwinia herbicola was used as a control [19].
The molecular weight of the levan polymer obtained was measured using multi-angle laser light-scattering gel permeation chromatography (MALLS–GPC). The chromatography system was equipped with an Ultrahydrogel Linear (0.78 × 30 cm, Waters) column, and analysis was done at 25 °C. The mobile phase contained 0.1 M of sodium nitrate (NaNO3) in 2% (v/v) acetic acid solution in water and was supplied to the system with a flow rate of 1.0 mL/min [19].
Biological Characterization
In-Vitro Antioxidant Activity
The antioxidant activity of obtained levan in this study was measured in vitro by free radical 2,2-Diphenyl-1-picrylhydrazyl (DPPH) scavenging using a modified approach published by [37]. Purple-colored DPPH is known to react with antioxidant compounds to form a colorless product of 1,1-diphenyl-2-picrylhydarzine (from dark purple to light yellow) and the reaction was analyzed by absorbance decrease in 517 nm. DPPH was dissolved in methanol to obtain a 1 mM concentration. Levan test solutions were prepared by dissolving in water and diluting in methanol to obtain (serial dilution between 1000 and 31.25 µg/mL). Each well received a 50 µL mM DPPH solution and was incubated for 30 min in the darkness at room temperature. Absorbance was recorded at 517 nm. A mixture of DPPH and methanol was used as control, mixtures of samples and methanol were used as blank and ascorbic acid in similar concentrations with samples used as a positive control. Inhibition (%) was calculated using the following Eq. 2.
In Vitro Anti-Inflammatory Activity
The anti-inflammatory activity of the obtained levan was determined by the modified inhibition of the protein denaturation method [38]. 500 µL of varying concentrations of levan solution were combined with 450 µL of 5% (w/v) BSA solution. With 1 N HCl, the pH of the reaction media was set to 6.3. After cooling, 2.5 mL of PBS was added, and the samples were incubated at 37 °C for 30 min and 57 °C for 3 min. The absorbances of samples were measured at 416 nm and inhibition (%) was calculated using Eq. 2, diclofenac sodium as a standard.
In Vitro Cell Proliferation Test (WST-1)
To estimate the cell viability of pure levan obtained was tested with L929 Cell Line (ATCC CCL-1) at different concentrations (0, 62.5, 125, 250, 500, and 1000 µg/mL) for 24 and 48 h. The viability of the cells was investigated with WST-1 (4-[3-(4-iodophenyl)-2-(4-nitrophenyl)-2 H-5-tetrazolio]-1,3-benzenedisulfonate) (Roche Applied Science, Penzberg, Germany) cell proliferation kit. Briefly, cells at the 70% confluency were trypsinized and seeded onto a 96-well plate at the cell/well density of 1 × 104. After overnight attachment, the medium (DMEM complete with 10% FBS and 1% penicillin-streptomycin) (PAN Biotech, Aidenbach, Germany) was replaced with an experimental medium that contained purified levan from the halophilic producer in this study and incubated for 24 and 48 h at 37 °C in humidified air containing 5% CO2. After the incubation period, the WST-1 reagent was added to wells and incubated at 37 °C for two more hours at dark in 5% CO2. The absorbance was measured at 450 nm by Gen5 Biotek Microplate Reader (BioTek, Epoch, ABD). Untreated cells (cells on wells without any sample but DMEM media) were used as a control and considered 100% viable. All experiments were performed in triplicate and statistical analysis of in vitro test data was performed via GraphPad Prism 9 with One-Way ANOVA and Tukey tests. Data were presented as a mean of 95% confidence interval (CI). AP-value below 0.05 was considered statistically significant [19].
Results and Discussion
Levansucrase Gene Amplification and Relative Quantification Real-Time PCR
After the amplification of the levansucrase gene region of the 87 isolates, sequencing, and alignment were performed. Seventeen isolates were selected for further molecular analysis, and RNA was extracted from these isolates for cDNA synthesis. The 16 S rRNA gene region was used as the reference gene, and the relative quantity of this region was determined for each organism. The levansucrase gene expression was then studied for each organism, and Ct values were calculated using the equation of 2−ΔΔCT (Table 1). P19-1 and P29-1 were found to be the microorganisms with the highest relative quantity. Levan production experiments were set up for all organisms based on these results. Q-PCR was suggested as a new method for the screening of genes directly related to the production of enzyme-based biopolymers. The primary advantages of Q-PCR are that it allows for the rapid and high-throughput detection and quantification of target DNA sequences in a variety of matrices. Simultaneous amplification and viewing of newly produced DNA amplicons help to reduce amplification time [39]. Additionally, Q-PCR can be used to identify target DNA and demonstrate the presence of specific genes and/or alleles [40]. Therefore, Q-PCR could be useful in showing the presence gene/genes responsible for enzyme-based polymer synthesis.
The findings of this study demonstrate the potential of Q-PCR for identifying microorganisms with high levan production potential and highlight the importance of gene expression studies in predicting levan production capabilities.
According to the gene expression analysis, the highest expression level was observed in the isolate coded P19-1, which was analyzed by 16 S rRNA amplification [41]. This strain was identified as Chromohalobacter salexigens P19-1 based on 16 S rRNA sequence analysis.
Validation of Levan Yields
Screening of isolates grown in a levan polymer medium for levan production was also carried out with the colorimetric method. Levan quantification was performed using the sugar determination method and fructose standard curve, and the obtained values are presented in Table 1. Interestingly, some microorganisms, such as P23-1, P23-2, P3-A, and P29-3 strains, produced high amounts of levan despite having low or no relative quantification values for levansucrase gene expression. These findings suggest that the expression of the levansucrase gene may not be the only factor influencing levan production in these microorganisms. Further studies are needed to elucidate the molecular mechanisms underlying levan production in these strains and to identify other potential genes involved in this process.
However, we now suggest a new hypothesis that different pathways may be active in levan production, and/or gene expression may be influenced by short-term stresses. Previous research by Ates and colleagues identified key network elements, such as enzymes and metabolites, involved in the mechanism of levan biosynthesis. They constructed an in silico metabolic model and analyzed it to identify critical components. According to their results, they identified three metabolic pathways related to levan biosynthesis: glycolysis, the pentose-phosphate pathway, and fructose-mannose metabolism. They also found three points – ribulose 5-phosphate, glucose 6-phosphate, and mannitol – at the branches of these pathways as critical components [42]. All of the microorganisms tested in this study were found to produce levan, suggesting that different pathways may be more effective than levansucrase expression for this biosynthesis.
Genome Assembly and Annotation Details
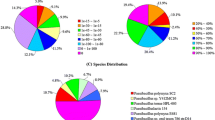
The reads obtained from sequencing were quality controlled with FASTQc and showed they were of good quality. The complete genome of Chromohalobacter salexigens P19-1 was assembled into 15 contigs, annotated, and found to consist of 3,711,318 bp base pairs, with an average G + C content of 63.94%. The genome organization exhibited a typical circular structure, characteristic of most bacterial genomes (Fig. 1A). The overall genome size and organization of C. salexigens P19-1 provide a fundamental framework for understanding its genetic potential and evolutionary history. The details of other statistics are given in Table 2 below.
The COG table (Table 3) summarizes the abundance and functional classification of genes based on their predicted protein products. A total of 3222 genes were assigned to different COG categories, providing insights into the genomic features and potential biological functions of the bacterial strain. The COG analysis revealed a diverse distribution of genes across various functional categories. The most abundant category was represented by COG code S, with 598 genes (16.66% of the total), indicating a lack of functional prediction for a significant portion of the genes. The next most prevalent category was COG code E, comprising 340 genes (9.47%), associated with amino acid metabolism and transport. Lastly, 127 genes (3.54%) did not fall within any of the defined COG categories, as indicated by the “-” code, suggesting their potentially unique or uncharacterized functions. Overall, the COG analysis provided valuable insights into the genomic composition and functional repertoire of the bacterial strain under investigation. The distribution of genes across various COG categories sheds light on the potential biological processes and molecular mechanisms driving the bacterial genome’s functional diversity.
Carbohydrate Metabolism and Energy Generation
C. salexigens P19-1 possesses the genes to synthesize levan from sucrose using the levansucrase gene. The gene sequence was submitted to NCBI blast for further investigation. Blast results revealed its homology to the levansucrase genes of Halomonas sp. THAF12 (88.41%) and Halomonas smyrnensis AAD6 (88%).
Subsystems annotation provided valuable insights into carbohydrate metabolism which contained 174 genes (Fig. 1B). Of these, 73 genes were associated with central carbohydrate metabolism, suggesting that the organism possesses a robust set of enzymes involved in the breakdown and utilization of carbohydrates. This also suggests that the organism can efficiently derive energy from carbohydrate sources.
The annotation results also highlight the presence of specific metabolic pathways involved in various processes such as methylglyoxal metabolism, pyruvate metabolism, glyoxylate bypass, TCA cycle, pentose phosphate pathway, and organic acid metabolism. These pathways play essential roles in energy production, carbon metabolism, and biosynthesis of important cellular components.
The identification of 24 genes associated with fermentation indicates that the organism can utilize alternative metabolic pathways under anaerobic conditions. Fermentation pathways provide a way to generate energy in the absence of oxygen and may be important for the organism’s survival in certain environments.
Also, the annotation results reveal the presence of genes related to the metabolism of specific sugars such as monosaccharides, mannose, D-gluconate, xylose, and L-fucose. This suggests that the organism can utilize these sugars as carbon sources for growth and metabolism.
Lastly, genes involved in the metabolism of organic acids, sugar alcohols, and other organic compounds were annotated suggesting a diverse metabolic capacity, allowing it to utilize a wide range of organic substrates as energy sources. Knowledge of carbohydrate metabolism is necessary for understanding possible levan production capabilities in different environments. It also helps to provide insights into its ecological niche, interactions with the environment, and potential applications in biotechnology and bioprocessing.
Nitrogen Metabolism
Further exploration of the annotated genome revealed important insights into the organism’s nitrogen metabolism (Fig. 1B). C. salexigens P19-1 possesses genes for converting nitrate and nitrite into ammonia (3) and assimilating ammonia (7). Saline soils often pose challenges to plant growth due to high salt concentrations, which can disrupt nutrient availability, including nitrogen [43]. By effectively utilizing and processing nitrogen compounds, the organism can contribute to nitrogen cycling in the soil, potentially enhancing nitrogen availability for plants.
Secondary Metabolism and Metabolite Gene Clusters
Subsystems annotation revealed 4 genes associated with auxin biosynthesis. Auxin biosynthesis, specifically in the context of bacterial secondary metabolism, refers to the production of auxin or auxin-like compounds by bacteria. Auxins are a class of plant hormones that regulate various aspects of plant growth and development. They are primarily known for their role in promoting cell elongation, root development, and tropical responses. However, recent research has revealed that certain bacteria are capable of synthesizing auxin or auxin-like compounds, which can influence plant growth and development [44]. Auxins may serve multiple purposes, including Plant growth promotion, plant-microbe interactions, biocontrol and disease suppression, and environmental adaptation. It is However important to note that the exact mechanisms and functions of auxin biosynthesis in bacterial secondary metabolism can vary depending on the bacterial species and the specific ecological context. C. salexigens P19-1 has been described in other studies as a plant-growth-promoting bacteria in saline habitats with studies citing ectoine as the metabolite promoting plant growth [45]. The ability to synthesize auxin may provide a novel mechanism through which C. salexigens P19-1 interacts with plants and survives high salinity conditions. Further research is nevertheless needed to elucidate the specific roles of auxin biosynthesis in C. salexigens P19-1 secondary metabolism.
The presence of secondary metabolite gene clusters was investigated using the antiSMASH software. This analysis identified several putative gene clusters associated with the NI-siderophore, and two RIPP-like clusters. The presence of these gene clusters suggests the potential for NI-siderophore, and two RIPP-like clusters to produce diverse secondary metabolites with biological activities. NI-siderophore is a type of siderophore, which is a small molecule secreted by bacteria to scavenge iron from the environment. Siderophores are essential for bacterial growth and survival, as iron is a critical nutrient required for various cellular processes. NI-siderophore specifically refers to a class of siderophores produced by certain bacteria.
RIPP-like clusters, on the other hand, refer to gene clusters that encode for Ribosomally Synthesized and Post-translationally Modified Peptides (RIPP). These clusters contain genes responsible for the biosynthesis and modification of RIPP-like peptides. RIPP-like peptides are a class of small, bioactive peptides that are ribosomally synthesized and undergo post-translational modifications to attain their final structure and function. They often possess antimicrobial or cytotoxic activities and can be produced by diverse microorganisms, including bacteria, fungi, and plants.
Phylogenetic Classification
The phylogenetic tree was built using representative genomes from NCBI. These genomes were identified using the Mash/MinHash algorithm [46]. The phylogenetic position of the genome was determined by selecting PATRIC global protein families [47], aligning them with MUSCLE [48], and their nucleotides mapped to the protein alignments. These were then arranged into a matrix that was then analyzed by bootstrapping with RaxML [49]. The results show that the genome under study is closest to C. salexigens DSM 3043 (Fig. 1C).
Optimization by Response Surface Methodology
Preliminary experiments conducted in this study showed that carbon source concentration, agitation rate, and incubation time had a significant effect on levan production. The maximum levan yield was obtained at a 1.0 vvm aeration rate in the fermenter, and this aeration rate was kept constant during the optimization studies. Response surface methodology was used to determine the optimum levels of the process parameters, including carbon source concentration, agitation rate, and incubation time. Based on the results of single-factor experiments, the levels of the process variables were set as carbon source concentration of 30–60 g/L, agitation rate of 150–450 rpm, and incubation time of 24–72 h for CCD experiments. After each fermentation, levan yield was measured, and the results are presented in Table S1 (see supplementary information). The results of CCD based on levan yields were statistically analyzed using ANOVA (Table S2) (see supplementary information).
The analysis conducted in this study provides the value of the model and determines the requirement of a more complex model with a fit. According to the obtained results, the R2 value was 0.869, indicating that the model as fitted explained 86.9% of the variability in levan yield. The F test for regression was significant at a level of 5% (p < 0.05), indicating that the model can explain the variation observed in levan yield with the designed levels of the factors. As shown in Table 5, the lack of fit (0.1325) was not significant at the 5% level (p > 0.05), indicating that the experimental data obtained fit well with the model. This means that the effects of carbon source concentration, incubation time, and mixing speed on levan production can be well explained by the model created. Using the regression coefficients found as a result of the multiple regression analysis on the experimental data, the quadratic model equation for the face-centered design was produced and given below.
The study presents the regression coefficients of the terms in the model equation in Table S2. The significance of each coefficient is assessed using P values, where a smaller P value indicates a stronger correlation of the coefficient. The results indicate that the carbon source concentration has the most significant effect on the levan amount, as it has the highest linear coefficient (52.243). The incubation time (31.696) and agitation rate (0.871) follow in significance. Furthermore, the study finds that carbon source concentration, agitation rate, and incubation time have a negative quadratic effect on the response (P < 0.05). This suggests that the amount of levan increases with increasing values of carbon source concentration, agitation rate, and incubation time, but the amount of levan decreases as higher values continue (Figure S1) (see supplementary information).
To determine the optimal conditions for the levan yield process, the second-order polynomial equation was utilized. The equation was solved, and the optimal conditions were found to be a carbon source concentration of 45 g/L (X1), agitation rate of 300 rpm (X2), and incubation time of 48 h (X3). The model estimated that under these conditions, a maximum levan yield of 734.91 mg/L could be obtained at the end of the fermentation process. To test the accuracy of the model, levan production was carried out in the production medium containing C. salexigensis P19-1 bacteria and sucrose, using the optimal conditions determined by the model. The results showed that the maximum amount of levan obtained at the 48th hour of fermentation was 709.14 mg/L, which is very close to the predicted value of 734.91 mg/L.
Analytic Methods and Chemical Characterization
Sugar Profile Analyses of C. salexigensis P19-1 Levan
The HPLC analysis conducted on C. salexigensis P19-1 levan (ChL) revealed that fructose was the primary monosaccharide present, as illustrated in Figure S2 (see supplementary information). The chromatogram of the standard mixture displayed a major peak at t = 6.98 min, which corresponded to fructose. In comparison, the commercial Erwinia herbicola levan (EhL) exhibited a distinct fructose peak at t = 6.68 min. However, the chromatogram of ChL displayed a broad fructose peak at approximately t = 6.03 min. These findings suggest that ChL levan is a fructose polymer, with fructose being the primary monosaccharide constituent.
Chemical Characterization
The chemical characterization of levan was performed using FT-IR analysis, and the results are presented in Fig. 2. The distinctive peaks of the obtained levan sample and the commercial levan produced by E. herbicola are shown in the spectrum. The results indicate that the polymer obtained from the halophilic isolate is a levan-type polysaccharide, as evidenced by the similarity between the sample and the pure and commercial levan. The FTIR spectrum of the obtained levan sample and the commercial levan mostly overlap, with characteristic peaks indicating the presence of fructose residue. Specifically, the bands around 3369 cm− 1, 2945 cm− 1, and 2453 cm− 1 correspond to the vibrations of OH stretching of fructofuranose residues, -CH2OH group, and C-H stretching of fructofuranose residues, respectively. Additionally, bands in the 1430–1220 cm− 1 range were produced by C-H vibrations combined with aromatic skeletal vibrations. The glycosidic linkage of C-O-C vibrations of fructofuranose rings and glycosidic linkages at 1119 − 1054 and 952 cm− 1 were also observed. These findings are consistent with previous studies [8] and confirm that the polymer obtained from the halophilic isolate is a fructose-based levan-type polysaccharide. The spectral peaks of the two polymer structures exhibited significant overlap when compared to the nuclear magnetic resonance (NMR) spectrum of commercially available EhL, which had a purity of 99%. (Fig. 3).
ChL had a weighted average molecular weight (Mw) of 1.770 × 106±0.01 g/mol, the number average molecular weight (Mn) of 9.911 × 105±0.01 g/mol, and polydispersity (Mw/Mn) values were found to be 1.786 ± 0.02. Poli et al. reported that levan produced by H. smyrnensis AAD6T the weight average molecular weight (Mw) was found to be 1 × 106 ± 0.1 g/mol [8].
Biological Activities
The in vitro scavenging activity of ChL was assessed using a non-enzymatic 2,2-diphenyl-1-picrylhydrazyl (DPPH) assay. The concentration-dependent percentage inhibition results are presented in Figure S3 (see supplementary information). The experiments were conducted in triplicate, and the results were reported as the mean ± standard deviation (SD).
Ascorbic acid was used as a control and exhibited antioxidant activity ranging from 85 to 140% for doses between 12.5 and 500 µg/mL. ChL demonstrated antioxidant activity ranging from 23.40 to 64.01%. At a concentration of 500 µg/mL, ChL exhibited strong antioxidant activity of 64.01% and was the most effective radical scavenger. ChL demonstrated stronger antioxidant activity at a lower concentration than levan produced from Bacillus velezensis VTX20 [50] and was comparable to levan produced from Bacillus subtilis natto KB1 [37] at the same concentration.
To evaluate the anti-inflammatory efficacy of ChL, inhibition of protein denaturation was employed. Figure S4 displays the percentage inhibition results as a function of concentration (see supplementary information). Diclofenac sodium, a commonly used anti-inflammatory medication, exhibited 53.48–99.82% anti-inflammatory activity at concentrations of 50–250 µg/mL, while ChL demonstrated 17.38–71.52% anti-inflammatory activity at concentrations of 50–400 µg/mL. Anti-inflammatory compounds are in high demand in the pharmaceutical industry, and the ChL was found to exhibit significant anti-inflammatory activity in this study.
The results of the cell viability analysis of the L929 cell line after 24 and 48 h of incubation with ChL are presented in Figure S5 (see supplementary information). The WST-1 cell proliferation assay revealed that the cellular viability at the end of 24 h was 100%, 329.09%, 353.12%, 360.93%, 360.64%, and 372.17% for the control, 62.5, 125, 250, 500, and 1000 µg/mL doses of ChL samples, respectively. At the end of 48 h, the cellular viability was 100%, 239.43%, 232.57%, 232.71%, 221.20%, and 218.52% for the control, 62.5, 125, 250, 500, and 1000 µg/mL doses of ChL samples, respectively. ChL exhibited high cell viability for 24 and 48 h at all concentrations studied in the L929 cell line and did not have a cytotoxic effect. In a previous study by Erdal Altıntaş and colleagues, high viability was observed in another cell line, the human dermal fibroblast cell line (PCS-201-012), after treatment with purified levan from Halomonas elongata 153B for all tested concentrations and durations of 24, 48, and 72 h [19]. In the present study, higher viability was observed in the L929 cell line used to investigate purified levan at all doses.
Conclusion
We used Q-PCR to screen genes responsible for enzyme-based biopolymer synthesis and identified the best levan producer among 87 isolates by expressing the levansucrase gene. Q-PCR reduced the time and number of tests required, making it a promising method for gene-linked biopolymer production. The obtained ChL had antioxidant, anti-inflammatory, and non-cytotoxic properties. Whole genome analysis of C. salexigens P19-1 revealed its genotypic potential, including the levansucrase gene involved in levan biosynthesis and versatile carbohydrate metabolism. Nitrogen metabolism capabilities, auxin biosynthesis, and other secondary metabolites and clusters suggest additional biotechnological importance in supporting plant growth in hypersaline environments.
Data Availability
This Whole Genome Shotgun project has been deposited at GenBank under the accession JAUKTO000000000. The version described in this paper is version JAUKTO010000000.
References
Combie J, Toksoy Öner E (2023) Fructan Production Processes. In The Book of Fructans Elsevier. (pp. 187–199)
Lippmann EO (1881) Ueber das Lävulan, Eine Neue, in Der Melasse Der Rübenzuckerfabriken Vorkommende Gummiart. Ber Dtsch Chem Ges 14(1):1509–1512
Greig-Smith RG (1901) The gum fermentation of sugar cane juice. Proc Linn Soc 26:589–625
Tomulescu C, Stoica R, Sevcenco C, Căşărică A, Moscovici M, Vamanu A (2016) Levan-A mini review. Sci Bull Series F Biotechnologies XX:309–317
Öner ET (2023) Levan as a Functional Polymer for Biomedical Applications. In The Book of Fructans. Elsevier. (pp. 257–274)
Erkorkmaz BA, Kırtel O, Abaramak G, Nikerel E, Toksoy Öner E (2022) UV and chemically induced Halomonas smyrnensis mutants for enhanced levan productivity. J Biotechnol 356:19–29.
Hamid AA, Rahmi K (2015) Optimization of cultivation medium for levan production using Bacillus subtilis in semi-industrial scale. J Sci Ind Res 77:386–393.
Poli A, Kazak H, Gürleyendaǧ B, Tommonaro G, Pieretti G, Öner ET, Nicolaus B (2009) High level synthesis of levan by a novel Halomonas species growing on defined media. Carbohydr Polym 78(4):651–657
Kırtel O, Versluys M, Van Den Ende W, Toksoy Öner E (2018a) Fructans of the saline world. Biotechnol Adv 36(5):1524–1539
Hamada MA, Hassan RA, Abdou AM, Elsaba YM, Aloufi AS, Sonbol H, Korany SM (2022) Bio_fabricated levan polymer from Bacillus subtilis MZ292983. 1 with antibacterial, antibiofilm, and burn healing properties. App Sci 12(13):6413
Laddha NB, Chitanand MP (2017) Screening of Levan producing Bacteria from Soil Collected from Jaggery Field. Int J Pharm Chem Biol Sci 7(3):202–210
Krishna PS, Woodcock SD, Pfeilmeier S, Bornemann S, Zipfel C, Malone JG (2022) Pseudomonas syringae addresses distinct environmental challenges during plant infection through the coordinated deployment of polysaccharides. J Exp Bot 73(7):2206–2221
Gross M, Rudolph K (1987) Studies on the Extracellular Polysaccharides (EPS) produced in vitro by Pseudomonas Phaseolicola: II. Characterization of Levan, Alginate, and ‘LPS’. J Phytopathol 119(3):206–215
Ozimek LK, Kralj S, Van Der Maarel MJEC, Dijkhuizen L (2006) The levansucrase and inulosucrase enzymes of Lactobacillus reuteri 121 catalyse processive and non-processive transglycosylation reactions. Microbiology 152(4):1187–1196
Demir EK, Nural Yaman B, Aytar Çelik P, Puhakka JA, Sahinkaya E (2021) Simulated acid mine drainage treatment in iron oxidizing ceramic membrane bioreactor with subsequent co-precipitation of iron and arsenic. Water Res 201:117297
Pfaffl MW (2007) Relative quantification. In: Dorak M (ed) Real-time PCR. Taylor & Francis, pp 89–108
Kirtel O, Menéndez C, Versluys M, van den Ende W, Hernández L, Toksoy Öner E (2018) Levansucrase from Halomonas smyrnensis AAD6T: first halophilic GH-J clan enzyme recombinantly expressed, purified, and characterized. Appl Microbiol Biotechnol 102(21):9207–9220.
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆CT method. Methods 25(4):402–408.
Erdal Altıntaş Ö, Toksoy Öner E, Çabuk A, Aytar Çelik P (2023) Biosynthesis of Levan by Halomonas elongata 153B: optimization for enhanced production and potential Biological activities for Pharmaceutical Field. J Polym Environ 31(4):1440–1455.
Aytar Celik P, Barut D, Enuh BM, Erdogan Gover K, Yaman N, Mutlu BB, Cabuk M, A (2023) A novel higher polyhydroxybutyrate producer Halomonas halmophila 18H with unique cell factory attributes. Bioresour Technol 372:128669
Andrews S (2010) Babraham Bioinformatics - FastQC A Quality Control tool for High Throughput Sequence Data [WWW Document]. Braham Bioinforma. URL https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed 6.21.23).
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Olson RD, Assaf R, Brettin T, Conrad N, Cucinell C, Davis JJ, Dempsey DM, Dickerman A, Dietrich EM, Kenyon RW, Kuscuoglu M, Lefkowitz EJ, Lu J, Machi D, Macken C, Mao C, Niewiadomska A, Nguyen M, Olsen GJ, Overbeek JC, Parrello B, Parrello V, Porter JS, Pusch GD, Shukla M, Singh I, Stewart L, Tan G, Thomas C, VanOeffelen M, Vonstein V, Wallace ZS, Warren AS, Wattam AR, Xia F, Yoo H, Zhang Y, Zmasek CM, Scheuermann RH, Stevens RL (2023) Introducing the bacterial and Viral Bioinformatics Resource Center (BV-BRC): a resource combining PATRIC, IRD and ViPR. Nucleic Acids Res 51:D678–D689
Wick RR, Judd LM, Gorrie CL, Holt KE (2017) Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13(6):e1005595.
Gurevich A, Saveliev V, Vyahhi N, Tesler G (2013) QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075
Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA, Stevens R, Vonstein V, Wattam AR, Xia F (2015) RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365
Seemann T (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069
Cantalapiedra CP, Hernández-Plaza A, Letunic I, Bork P, Huerta-Cepas J (2021) eggNOG-mapper v2: functional annotation, Orthology assignments, and Domain Prediction at the Metagenomic Scale. Mol Biol Evol 38:5825–5829
Blin K, Shaw S, Augustijn HE, Reitz ZL, Biermann F, Alanjary M, Fetter A, Terlouw BR, Metcalf WW, Helfrich EJN, van Wezel GP, Medema MH, Weber T (2023) antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res 51(W1):W46–W50.
Kanehisa M, Goto S (2000) KEGG: Kyoto Encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35:W182–W185
Erdal Altıntaş Ö, Aytar Çelik P (2023) Levan-based bioactive hydrogels containing herbal liposome in treating acute wounds. J Bioact Compat Polym 38(6):437–457.
Viikari L (1984) Formation of levan and sorbitol from sucrose by Zymomonas mobilis. Appl Microbiol Biotechnol 19(4):252–255
Ghose TK (1987) Measurement of cellulase activities. Pure Appl Chem 59:257–268.
Goldman D, Lavid N, Schwartz A, Shoham G, Danino D, Shoham Y (2008) Two active forms of Zymomonas mobilis Levansucrase. J Biol Chem 283(47):32209–32217
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72(1–2):248–254
Domżał-Kędzia M, Lewińska A, Jaromin A, Weselski M, Pluskota R, Łukaszewicz M (2019) Fermentation parameters and conditions affecting levan production and its potential applications in cosmetics. Bioorg Chem 93:102787
Mummaleti G, Sarma C, Kalakandan SK, Gazula H, Sivanandham V, Anandharaj A (2022) Characterization of levan produced from coconut inflorescence sap using Bacillus subtilis and its application as a sweetener. LWT 154:112697
Kralik P, Ricchi M (2017) A basic guide to real time PCR in microbial diagnostics: definitions, parameters, and everything. Front Microbiol 8:108
Klein D (2002) Quantification using real-time PCR technology: applications and limitations. Trends Mol Med 8(6):257–260
Nural Yaman B, Aytar Çelik P, Mutlu MB, Çabuk A (2020) A combinational analysis of Acidophilic bacterial diversity of an Iron-Rich Environment. Geomicrobiol J 37(10):877–889.
Ates O, Arga KY, Oner ET (2013) The stimulatory effect of mannitol on levan biosynthesis: lessons from metabolic systems analysis of Halomonas smyrnensis AAD6T. Biotechnol Prog 29(6):1386–1397
Du X, Ge Y, Zhang, Yun, Hu H, Zhang, Yiying, Yang Z, Ren X, Hu S, Feng H, Song Y (2023) Responses of soil carbon cycling microbial functional genes to nitrogen and phosphorus addition in saline-sodic soils. Plant Soil 490:261–267
Etesami H, Glick BR (2020) Halotolerant plant growth–promoting bacteria: prospects for alleviating salinity stress in plants. Environ Exp Bot 178:104124
Srivastava AK, Srivastava R, Sharma A, Bharati AP, Yadav J, Singh AK, Tiwari PK, Srivatava AK, Chakdar H, Kashyap PL, Saxena AK (2022) Transcriptome analysis to understand salt stress regulation mechanism of Chromohalobacter salexigens ANJ207. Front Microbiol 13:909276
Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM (2016) Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 17:132
Davis JJ, Gerdes S, Olsen GJ, Olson R, Pusch GD, Shukla M, Vonstein V, Wattam AR, Yoo H (2016) PATtyFams: protein families for the Microbial genomes in the PATRIC Database. Front Microbiol 7:118
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Vu THN, Quach NT, Nguyen NA, Nguyen HT, Ngo CC, Nguyen TD, Ho P-H, Hoang H, Chu HH, Phi Q-T (2021) Genome Mining Associated with Analysis of structure, antioxidant activity reveals the potential production of Levan-Rich exopolysaccharides by Food-Derived Bacillus velezensis VTX20. Appl Sci 11(15):7055
Funding
This study was supported within the scope of TUBITAK 1001-The Scientific and Technological Research Projects Funding Program (Project Number: 220S761).
Open access funding provided by the Scientific and Technological Research Council of Türkiye (TÜBİTAK).
Author information
Authors and Affiliations
Contributions
Conceptualization: Aytar Celik P., Erdal Altıntaş Ö., Cabuk A., Toksoy Öner E., Mutlu MB., Nural Yaman B; Enuh BM.; methodology: Aytar Celik P., Barut D., Nural Yaman B., Erdal Altıntaş Ö., Enuh BM.; validation: Aytar Celik P., Toksoy Öner E., Mutlu MB., Cabuk A.; resources: Aytar Celik P., Cabuk A.; references: Nural Yaman B., Barut D.; writing-original draft preparation: all authors; writing-review and editing: Toksoy Oner E., Aytar Çelik P., Çabuk A.; visualization: Barut D., Erdal Altıntaş Ö., Nural Yaman B., Enuh BM.
Corresponding author
Ethics declarations
Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Aytar Çelik, P., Barut, D., Erdal Altintaş, Ö. et al. QPCR-Guided Screening of Levansucrase: Levan Characterization and Genomic Insights. J Polym Environ (2024). https://doi.org/10.1007/s10924-024-03197-6
Accepted:
Published:
DOI: https://doi.org/10.1007/s10924-024-03197-6