Abstract
In the rapidly evolving field of medical image analysis utilizing artificial intelligence (AI), the selection of appropriate computational models is critical for accurate diagnosis and patient care. This literature review provides a comprehensive comparison of vision transformers (ViTs) and convolutional neural networks (CNNs), the two leading techniques in the field of deep learning in medical imaging. We conducted a survey systematically. Particular attention was given to the robustness, computational efficiency, scalability, and accuracy of these models in handling complex medical datasets. The review incorporates findings from 36 studies and indicates a collective trend that transformer-based models, particularly ViTs, exhibit significant potential in diverse medical imaging tasks, showcasing superior performance when contrasted with conventional CNN models. Additionally, it is evident that pre-training is important for transformer applications. We expect this work to help researchers and practitioners select the most appropriate model for specific medical image analysis tasks, accounting for the current state of the art and future trends in the field.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
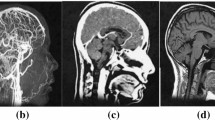
Convolutional neural networks (CNNs) are a type of deep learning algorithm and a key technology behind the modern field of artificial intelligence (AI) [1]. They consist of multiple convolutional layers with nonlinear activation functions, pooling layers, and fully connected layers, enabling them to capture complex patterns and textures in images, rendering them particularly well-suited for visual data interpretation applications [2]. Consequently, CNNs play a pivotal role in medical image analysis, extracting and improving the accuracy and efficiency of image-based medical applications. For instance, CNNs find application in the classification, segmentation, and registration of various medical images such as endoscopic, X-ray, magnetic resonance imaging (MRI), computed tomography (CT), ultrasound (US), skin, and histopathology images [3,4,5,6,7,8,9,10]. An example of the application of CNN to brain MRI is shown in Fig. 1. CNNs contribute to disease detection, including bone fractures, pneumonia, and cancer, as well as predicting cancer prognosis and pathological classification based on genetic mutations [11,12,13,14,15,16]. Moreover, a number of CNN-based segmentation models have been reported to exhibit performance comparable to that of human experts [17,18,19]. Despite the advanced capabilities of CNN in medical image analysis, they possess certain limitations. A primary concern is their lack of explainability; CNNs often operate as black boxes, providing minimal insights into how they reach conclusions, although several techniques, such as gradient-weighted class activation mapping (Grad-CAM), have been developed [20]. This opacity poses a significant challenge in clinical settings where understanding the decision-making process is crucial for diagnosis and treatment. Additionally, CNNs are prone to domain shift problems; for example, their performance may degrade when exposed to data differing from the training dataset, such as images from different medical centers or imaging devices [21,22,23]. This vulnerability raises concerns about the reliability and generalizability of these findings across different clinical environments.
An example of medical image analysis using CNN (brain MRI). CNNs have input layers, output layers, many hidden layers, millions of parameters, and the ability to train complex objects and patterns. The input layer subsamples the input given by the convolution and pooling process and applies the activation function (ReLU in this figure). All of these layers are partially connected hidden layers, with the last fully connected layer being the output layer. The output retains its original shape, which is close to the dimensions of the input image
Although other techniques have been employed, such as recurrent neural networks, convolution was the mainstream approach in the field of image processing [24] until very recently (in the 2020s). Transformers, originally developed for natural language processing (NLP), have revolutionized the field of deep learning due to their unique architectures based on self-attention mechanisms [25]. Vision transformers (ViTs) adopt this powerful framework for image processing [26]. An example is shown in Fig. 2. Unlike traditional convolutional approaches, ViTs treat an image as a sequence of patches and apply a transform model to these patches, enabling them to learn spatial hierarchies and relationships in the visual data [27, 28]. This approach has achieved remarkable success, offering an alternative to CNNs with potentially greater flexibility and the ability to handle more diverse and complex image datasets. ViTs and their derived instances achieved state-of-the-art (SOTA) performance on several benchmark datasets [29,30,31]. Active research has been conducted on adding explainability to ViTs, including the use of attention maps to visualize the features detected by ViTs [32,33,34]. Although ViTs have positive aspects, they face unique challenges. First, they require larger datasets for effective training compared to CNNs [35,36,37], which can be critical in situations with limited data, such as medical data. Second, ViTs often require additional computational resources, making their deployment challenging in resource-constrained environments [38,39,40]. Last, their relative novelty implies less established knowledge and best practices for their applications compared to those for CNNs.
An example of medical image analysis using ViT (same brain MRI as used in Fig. 1). ViT consists mainly of the encoder part of the transformer. First, one input image is divided into N (P, P) resolution patches (\(\:\mathcal{x}\in\:\:{\mathbb{R}}^{H\:\times\:W\:\times\:C\:}\)→ \(\:{\mathcal{x}}_{\varvec{p}}\in\:\:{\mathbb{R}}^{N\:\times\:({P}^{2}\bullet\:C)}\)), where (H, W) is the resolution of the original image, and C is the number of channels. Then, with matrix E, project each patch onto a vector of length (\(\:{P}^{2}\bullet\:C\)) to D dimensions to create the location information of the original patch (\(\:{\mathbf{E}}_{pos}\:\in\:\:{\mathbb{R}}^{(N+1)\:\times\:D}\)). Combine these data as input data to the Transformer Encoder (\(\:{z}_{0}=\:\left[{x}_{class};\:{x}_{p}^{1}\mathbf{E};\:{x}_{p}^{2}\mathbf{E};\:\bullet\:\bullet\:\bullet\:;\:{x}_{p}^{N}\mathbf{E}\right]\:+\:{\mathbf{E}}_{pos})\). The output of the Transformer-Encoder is further input to MLP to obtain task-specific output (Class 0: Benign, Class 1: Tumor, etc.)
While numerous new models based on convolution and attention mechanisms have been proposed, direct comparisons between these two architectures are relatively rare. This scarcity is likely due to the challenges involved in ensuring that learning conditions are strictly equivalent. Considering the above, our central research question is, “When building a machine learning model using medical images as input, which architecture should be used: CNNs or transformers?”
To accurately address this question, we pose several sub-questions:
-
SQ1: Are there tasks that are well-suited for each model?
-
SQ2: Are there appropriate image modality types for each model?
-
SQ3: What are the optimal learning conditions for each model?
-
SQ4: Which architecture demonstrates greater robustness?
To address these questions, we conducted a literature review. Figure 3 illustrates the central research question and sub-questions guiding our systematic review. The remainder of this paper is organized as follows: In the “Basic Concepts and Historical Overview” section, we briefly present the basic concepts and historical overview of convolution and attention. The “Methods” section describes the research methodology, and the “Results” section presents the selected papers. Finally, in the “Discussion” section, we discuss the selected papers and answer the research questions.
Basic Concepts and Historical Overview
Concept of Convolution
In this section, we describe the concept of convolution in the context of CNNs. Convolution is a mathematical operation in which a filter comprising kernels is applied to an input image to extract its features. The convolution operation at position (x, y) is defined as:
I denotes the input image, and K, a 2-dimensional kernel of size (a + 1, b + 1), represents the convolutional kernel; the convolution operation (denoted by ∗) at a position (x, y), where I (x + i, y + j) denotes the pixel value of the image at position (x + i, y + j); and K(i, j) denotes the corresponding kernel value. Figure 4 illustrates the convolution operation. The kernel glides over the image, and the sum is calculated at each position to effectively filter the image. This process captures local patterns, such as edges, textures, and shapes, which are critical for image recognition tasks. Moreover, the depth, size, and number of kernels are key hyperparameters in CNNs that determine the ability of the network to extract different levels of features, from simple to complex. Learning in CNNs involves updating the kernel values, which is the essence of CNNs, allowing them to effectively learn hierarchies of image features.
In this specific example, aaa and bbb are both equal to 2, as the kernel K is a 3 × 3 matrix (with indices ranging from 0 to 2). In the top part of the figure, the kernel K is positioned over the top-left corner of the input matrix I. Each element of the kernel is multiplied by the corresponding element of the input matrix, and the results are summed to produce a single value (5 in this case), which is placed in the top-left position of the output matrix. In the bottom part of the figure, the kernel K slides to the next position to the right on the input matrix I. The same element-wise multiplication and summation process is performed, resulting in a value of 0, which is placed in the corresponding position of the output matrix. This process is repeated across the entire input matrix to generate the full output matrix, capturing important features, such as edges and patterns, in the input data.
Brief Historical Overview of Convolution
The concept of convolution in neural networks dates back to the 1980s with the introduction of the neocognitron by Kunihiko Fukushima in 1980 [41]. Inspired by the visual cortex of animals, this model included components called “simple” cells (S cells) and “complex” cells (C cells), laying the foundation for feature extraction through layered convolutions. In 1998, LeCun’s study marked an important milestone in the practical application of CNNs [42]. LeCun et al. demonstrated the effectiveness of CNNs in image-recognition tasks, particularly in the recognition of handwritten digits, using the LeNet-5 architecture. The popularity and utility of CNNs surged with the advent of deep learning and increased computing power in the 21st century [43]. The 2012 ImageNet Challenge (ISVRC-2012) was a pivotal moment where Krizhevsky’s AlexNet model significantly outperformed traditional image recognition methods [44]. This success showcased the power of deep CNNs in handling large-scale visual data and led to the rapid proliferation of CNN applications in various fields, particularly image and video recognition. Considerable progress has been made in the medical imaging applications of CNNs, including image anomaly detection, radiological image segmentation, and pathological slide analyses (Fig. 1) [7, 45,46,47,48]. The historical journey of CNNs, from their inception to their current status as the cornerstone of image analysis, highlights their transformative impact in both technology and healthcare fields.
Advantages, Disadvantages, and Applications of Convolution
CNNs effectively capture local patterns, such as edges, textures, and shapes, within images. This local feature extraction is critical for tasks where detailed spatial hierarchies are important. A key advantage of CNNs is parameter sharing, where convolutional kernels are shared across different regions of the image [44]. This reduces the number of parameters, making the model less complex and easier to train. In addition, convolution operations can be efficiently parallelized, leading to faster computations on modern GPUs. However, CNNs have difficulty capturing long-range dependencies due to their localized receptive fields. Fixed kernel sizes limit the ability to detect features at different scales and in different contexts within the image. Despite these limitations, CNNs are widely used for image recognition tasks, such as object detection, face recognition, and image classification (e.g., the ResNet and VGG architectures). They are also widely used in medical image analysis, including the analysis of MRI and CT scans for disease detection and diagnosis, including the identification of tumors or anomalies.
Concept of Attention
In deep learning, the attention mechanism, which is central to the structure of ViTs, signifies a paradigm shift from the localized receptive fields of CNNs. Vaswani et al. introduced attention in their seminal paper, “Attention is all you need.” The essence of the attention mechanism is to focus on different parts of the input data and dynamically weigh their importance [25]. The core concept can be encapsulated in the formula for the scaled dot product of attention as follows:
where Q, K, and V denote the query, key, and value matrices, respectively, derived from the input data; dk denotes the scaling factor; and dk is the key dimension. Figure 5 illustrates the attention operation. The softmax function can be applied to the scaled dot products of queries with keys to provide a distribution of weights [49]. These weights can then be applied to the values, resulting in an output that reflects the focused areas of the input. This attention mechanism enables the model to consider the entire input sequence globally, making it particularly adept at capturing long-range dependencies in the data. In the context of medical imaging, ViTs can detect patterns and correlations across an entire image, potentially providing a more holistic and detailed understanding compared to that by the localized approach of CNNs.
-
1.
Query (Q): Represents the set of vectors that will be compared against the key vectors to calculate attention scores.
-
2.
Key (K): Represents the set of vectors that the queries are compared to.
-
3.
Value (V): Represents the set of vectors that are weighted by the attention scores and combined to produce the output.
The attention scores were calculated using the scaled dot-product attention formula:
where dk is the dimension of the key vectors. The softmax function was applied to the scaled dot products of the query and key vectors to obtain the attention weights, which indicate the importance of each value vector in producing the final output. These weights were then used to combine the value vectors, resulting in the output vectors. The final output matrix was obtained by applying these attention-weighted value vectors.
Brief Historical Overview of Attention
The concept of attention in deep learning was initially introduced to address the limitations of sequential data processing, particularly in NLP. A seminal study marked the beginning of the evolution of attention mechanisms [50], demonstrating how attention allows a model to focus on different parts of the input sequence while generating each word of the output sequence, thereby improving the performance of machine translation systems. However, significant advancements occurred in 2017, as mentioned above [25]. This study introduced a transformer model that relies entirely on the attention mechanism and omits the recurrent layer commonly used in NLP tasks. The main concept introduced in this study was self-attention, which enables the importance of different parts of input data to be weighted relative to each other, thereby effectively capturing long-range dependencies. The introduction of transformers has marked a paradigm shift in deep learning. Originally designed for NLP, their architectures have proven highly effective for various tasks. The adaptability of the attention mechanism has led to its integration into various domains, including computer vision. The ViT model, introduced by Dosovitskiy et al. in 2020, adapts the transformer architecture to image classification tasks [26]. By treating an image as a sequence of patches and applying a self-attention mechanism, ViTs demonstrated remarkable performance, challenging the dominance of CNNs in image-related tasks. However, in medical imaging, the application of attention-based models is still in its infancy compared to that of CNNs. Nonetheless, early results are promising, particularly in tasks that require the analysis of large-scale patterns and contextual information in images (such as detecting anomalies in radiological scans or identifying patterns on histopathology slides) (Fig. 2) [49, 51, 52].
Advantages, Disadvantages, and Applications of Attention
Attention mechanisms consider the entire input sequence, allowing them to effectively capture long-range dependencies and global context. As they can process entire sequences in parallel, they are efficient and scalable. They use a flexible weighting scheme, where each part of the input can be dynamically assigned different levels of importance, improving the focus on relevant features [25]. However, computing attention weights for all input pairs leads to higher computational complexity and resource requirements. These models also require large amounts of data for training and are prone to overfitting when applied to smaller datasets. Attention mechanisms are central to NLP tasks, such as machine translation, question answering, and document summarization, as demonstrated by models such as BERT and GPT. They are also used in image processing tasks, such as image generation and captioning; for example, ViTs and SWIN transformer have shown promising results in capturing complex visual patterns [53].
Methods
The methodology utilized for conducting the literature review followed previously reported guidelines [54]. Google Scholar was used as the data source for extracting primary studies. The search strings used in the study were ‘((ViT) AND (CNN) AND (Medical Images) OR (Comparing) OR (Vision Transformers) OR (convolutional neural networks) OR (differences))’. The search was conducted in October 2023.
Inclusion Criteria
The inclusion criteria for selecting papers were that they had to be written in English and published between January 2021 and October 2023. This timeframe was chosen because ViTs were not proposed until the end of 2020 [26]. In addition, the studies had to compare CNNs and ViTs on medical images using any pre-trained model of the two architectures. Studies proposing a hybrid architecture combining the two architectures into one, with their results compared, were also considered. The dataset used in the studies was not specific but had to be an image dataset suitable for classification using both deep learning architectures. Studies validated using externally independent datasets were preferred; however, those validated using a single dataset were also included.
Exclusion Criteria
Studies exclusively focusing on one of the two deep learning architectures (i.e., ViTs or CNNs) were excluded. Another exclusion criterion was that papers with fewer than three citations were not considered.
Results
Search Results
In this study, 9,100 search results were obtained using the search strings. Of these, 52 papers met the inclusion criteria, and 16 of these were excluded; accordingly, 36 papers were included in analyses (Fig. 3). While our initial criteria included a wide range of tasks, including detection, reconstruction, survival analysis and prediction, video-based applications, and image synthesis, no studies met our criteria within these specific tasks. An overview of the 36 included studies is presented in Tables 1 and 2. Figure 6A shows the distribution of task categories across 36 eligible studies. The most prevalent task was classification, followed by segmentation and registration. Figure 6B shows the distribution of image modalities across 36 eligible studies. Radiography was the most commonly used imaging modality, followed by pathological imaging, MRI, and fundus imaging. Figure 6C displays the results indicating which technique (convolution or attention) was deemed the most effective. It is important to note that these results were based on the authors’ descriptions and were therefore subjective. Additionally, validation with an independent external text dataset was performed in only two of the papers.
Classification
Classification emerged as one of the most researched topics, with X-ray imaging being the most commonly used imaging type; hence, this combination was the most common, featured in ten papers. The popularity of X-ray imaging can be attributed to the ready availability of X-ray imaging data and numerous public X-ray imaging datasets. For instance, Nafisah et al., Usman et al., Tyagi et al., Chetoui et al., Okolo et al., and Murphy et al. attempted to detect pneumonia [33, 55, 60, 64, 68, 83]. Among these studies, the most noteworthy is that of Murphy et al. who utilized an independent external text dataset to rigorously compare the two architectures [83]. This study evaluated the target model performance, sampling efficiency, and hidden layer stratification in the analysis of chest and extremity radiographs. The key findings indicated that while CNNs, especially DenseNet121, slightly outperformed the data-efficient image transformer (DeiT)-B ViT in terms of diagnostic accuracy, both models demonstrated comparable sample efficiencies. Considerably, the ViT model exhibited reduced susceptibility to hidden stratification, a phenomenon in machine learning models in which predictions are made based on features not directly related to the condition or disease being diagnosed but rather on incidental, non-disease features present in the data. For example, in the context of disease classification using X-ray imaging, a model might erroneously associate the presence of a medical device, such as a chest tube, with a particular disease, such as pneumothorax. This misunderstanding is not because the medical device is an actual indicator of the disease but because the model has learned to correlate the presence of the device with the disease due to biases in the training data. Murphy et al. reported that the ViT model exhibited a lower tendency for hidden stratification than did the CNN model, suggesting that ViT is less susceptible to being misled by incidental features in medical images, potentially leading to more accurate and reliable disease classification.
Wu et al. conducted a study using an independent test dataset [57]. These findings demonstrate the potential of ViT in medical imaging, specifically for classifying emphysema subtypes. This study was notable for its ability to classify centrilobular, panlobular, and paraseptal emphysema from CT images, outperforming CNNs such as AlexNet, Inception-V3, and ResNet50 in terms of accuracy. A key aspect of this study was the use of a private dataset for training and a public dataset for testing. The ViT model achieved average accuracies of 95.95% and 72.14% for the private and public datasets, respectively, thus outperforming each CNN model. Moreover, the results were particularly good for the private test data and public test data, achieving average accuracies of 72.14% (ViT) vs. 66.07% (AlexNet, the best performance among the CNNs). This research highlighted the efficiency of ViT in handling data, requiring fewer images for training than do CNN models. Additionally, the study examined the interpretability of the model using attention rollout heat maps to visualize how ViT discriminated between different lung regions to classify emphysema types.
Fundus images are a relatively popular image type. Using the Kaggle diabetic retinopathy detection dataset [92], which poses challenges due to class imbalances, Wu et al. used data augmentation techniques (such as panning and rotating images) to enhance model training [80]. The methodology involved dividing fundus images into non-overlapping patches, linearly and positionally embedding them, and processing them through multihead attention layers in the ViT model. This approach yielded excellent results, with the model achieving an accuracy of 91.4%, specificity of 97.7%, precision of 92.8%, sensitivity of 92.6%, quadratic weighted kappa score of 0.935, and area under the curve (AUC) of 0.986, outperforming CNN models.
Deininger et al. compared the effectiveness of ViTs and CNNs in the field of digital pathology [56]. This study focused on the application of ViTs for tumor detection and tissue-type identification in whole-slide images (WSIs) of four different tissue types. The patchwise classification performance of the ViT model DeiT-Tiny was compared with that of the state-of-the-art (SOTA) CNN model ResNet18. Due to the limited availability of annotated WSIs, both models were trained on large volumes of unlabeled WSIs using self-supervised methods. The results showed that the ViT model slightly outperformed the ResNet18 model in tumor detection for three of the four tissue types, while the ResNet18 model performed slightly better in the remaining tasks. The aggregated predictions of both models correlated at the slide level, suggesting that they captured similar image features. Overall, the ViT model performed comparably to the ResNet18 model but required more training effort.
Pachetti et al. conducted a study using MRI-based images [75]. This study introduced and evaluated a modified 3D ViT architecture trained from scratch on the ProstateX-2 Challenge dataset. This study aimed to determine whether 3D ViTs could effectively predict the aggressiveness of prostate cancer based on the Gleason score, which has been diagnosed in a more invasive way. A key aspect of this research was a comparison of the performance of 3D ViT against a 3D CNN trained from scratch. The results showed that 3D ViT not only had the ability to predict cancer aggressiveness but also outperformed the 3D CNN in this task. However, the SOTA 2D CNN model—that is, the fine-tuned AlexNet model—outperformed the 3D ViT.
Gheflati and Rivaz presented a study on the use of ViT and CNN for breast US image classification [82]. Using two datasets with 943 breast US images, different pre-trained ViT models were compared with SOTA CNNs (ResNet50, VGG16, and NASNET models). A key finding was that ViTs, particularly the B/32 model, achieved high classification accuracy and AUC values that surpassed or matched those of the best CNN models. For example, the B/32 model achieved an accuracy of 86.7% and AUC of 0.95, demonstrating the potential of ViTs to efficiently process spatial information in medical images. An important aspect of this study was the demonstration that ViTs could achieve high performance even with smaller datasets, which is a considerable advantage in medical imaging, where large datasets are not always available.
The SkinTrans model proposed by Xin et al. was designed to focus on the most important features of skin cancer images while minimizing noise through a combination of multi-scale image processing and contrastive learning [59]. Two datasets were used for validation: the publicly available HAM10000 dataset, comprising 10,015 images from seven skin cancer classes, and a clinical dataset collected through dermoscopy, comprising 1,016 images, including three typical types of skin cancer. The SkinTrans model exhibited impressive results, achieving 94.3% accuracy on the HAM10000 dataset and 94.1% accuracy on the clinical dataset. The addition of the simple ViT model also achieved high accuracies of 93.5% and 93.4% on the respective datasets, outperforming CNN models. A notable aspect of this study was the use of Grad-CAM visual analysis, demonstrating that the proposed model could identify the most relevant areas in skin cancer images, indicating that it learned the correct features for accurate classification.
Segmentation
Segmentation was the second most popular task and is expected to have many practical applications. Swin-Unet, proposed by Cao et al. is a new purely transducer-based segmentation model (Fig. 7) [91]. For example, Swin-Unet is characterized by using non-overlapping image patches as tokens, processed through a transformer-based encoder-decoder structure with skip links. This design facilitates effective learning of local and global semantic features. Studies have demonstrated the superior performance of Swin-Unet in segmentation tasks on multiple datasets, including multiorgan and cardiac segmentations, highlighting its excellent accuracy [91]. Figure 8 shows an illustrative example of segmentation predictions by U-Net and Swin-Unet, both trained on the UW-Madison GI Tract Image Segmentation dataset, using the validation portion of the same dataset for reference [93, 94]. Hatamizadeh et al. proposed the Swin-UNEt TRansformer (Swin UNETR), which uses Swin transformers as encoders in a U-shaped network connected to a multiresolution CNN-based decoder via skip links [90]. Swin UNETR is characterized by its ability to learn multi-scale contextual information and model long-range dependencies, outperforming previous methods in Brain Tumor Segmentation (BraTS) 2021, a brain tumor segmentation challenge [95].
Gulzar and Khan presented an in-depth comparison of ViTs and CNNs for skin lesion segmentation in medical images [89]. This research is crucial for evaluating the effectiveness of these technologies in medical image analysis, particularly in the challenging areas of skin lesion detection and segmentation. Using the ISIC 2018 dataset [96], different architectures, including the U-Net, V-Net, Attention U-Net, TransUNet, and Swin-Unet models, were examined, and their performance in accurately segmenting skin lesions was evaluated. The results highlighted that the hybrid models, particularly TransUNet, exhibited superior performance in terms of accuracy and the Dice coefficient, outperforming other benchmarking methods. This study highlighted the potential benefits of integrating ViTs with traditional CNNs in medical imaging and demonstrated their effectiveness in handling complex tasks, such as skin lesion segmentation.
Example of medical image segmentation using Swin-Unet (lung CT image). The architecture was adapted from Cao et al. [91]. Swin-Unet consists of encoder, bottleneck, decoder, and skip connections. In the encoder, to convert the input to sequence embedding, a Linear Embedding layer is applied to project the feature dimension into an arbitrary dimension (represented as C) after the lung CT image is divided into 4 × 4 sized patches. The transformed patch token passes through several Swin Transformer Blocks and Patch Merging layers to produce a hierarchical feature representation. The decoder consists of Swin Transformer Blocks and Patch Expanding Layers. The extracted context features are fused with the multiscale features from the encoder via skip connections to complement the loss of spatial information due to down-sampling. The Patch Expanding layer reshapes the feature maps in adjacent dimensions into a larger feature map with twice the resolution up-sampled. The last Patch Expanding layer is used to perform 4 x up-sampling to restore the feature map resolution to the input resolution (W x H), and then a linear projection layer is applied to these up-sampled features to output pixel-level segmentation predictions
Comparison of segmentation results using U-Net and Swin-Unet on the UW-Madison GI Tract Image Segmentation dataset. The first row shows the ground truth annotations for the large bowel (red), small bowel (green), and stomach (blue). The second row presents the segmentation results from U-Net, trained for 100 epochs using the Adam optimizer with a learning rate of 2e-3, weight decay of 1e-6, and CosineAnnealingLR scheduler. The third row shows the segmentation results from Swin-Unet, trained for 100 epochs using the SGD optimizer with a learning rate of 0.05, momentum of 0.9, weight decay of 1e-4, and PolynomialDecayLR scheduler
Registration
Mok et al. proposed a unique application of transformer architecture to medical imaging [85]. In various medical imaging studies, rigid and affine registrations play a crucial role. The authors proposed a method named Coarse-to-Fine Vision Transformer (C2FViT) for 3D affine medical image registration (Fig. 9). Unlike traditional CNN-based techniques, this method demonstrated enhanced accuracy, robustness, and speed, particularly in scenarios with significant initial misalignment.
Overview diagram of C2FViT. This figure was prepared with reference to Mok et al. [85]. C2FViT uses convolutional patch embedding instead of the linear patch embedding approach. Their method had been divided into L stages that solved the affine registration with an image pyramid. All stages share the same architecture consisting of a convolutional patch embedding layer and Ni transformer encoder blocks. C2FViT solves the affine registration problem in a coarse-to-fine manner, and the intermediate input video Mi is transformed by progressive spatial transformation. Finally, the estimated affine matrix AL in the final stage is employed as the output of the model fθ. In this figure, L is three
Discussion
Although we identified many studies that used attention, convolution, or a combination of both, few studies have directly compared these two approaches. Studies that do not simply cite results from other papers as benchmarks but instead run multiple models using the same dataset for comparison are particularly valuable. In addition, most studies rely solely on publicly available datasets, and few papers conducted comparisons using multiple datasets, including private datasets. As a result, it is difficult to fully answer our original research question and sub-questions. Nevertheless, we attempted to address these questions based on the available data. Figure 10 provides a visual summary of the central research question and SQs addressed in the systematic review, along with their corresponding answers.
SQ1: Are There Tasks that are well Suited for each Model?
Although numerous studies suggest that attention mechanisms generally outperform convolution methods, our discussion focuses on studies that highlight scenarios where convolution is superior [65, 73, 75]. The summarized results are presented in Table 3. It is evident that tasks in which convolution excels are exclusively related to classification [65, 73, 75]. Given that there is only one example of registration, it is premature to draw definitive conclusions. In contrast, for segmentation tasks, pure attention models or those combining attention and convolution models tend to outperform other models. This superiority may stem from the ability of attention mechanisms to capture long-range dependencies and enable attention-based models to integrate information across the entire image [26, 97]. Consequently, incorporating attention mechanisms into the design of segmentation models may prove beneficial for achieving improved results.
SQ2: Are There Appropriate Image Modality Types for each Model?
Upon examining datasets from studies asserting the superiority of convolution, no common characteristics emerged. Therefore, switching mechanisms based on the type of image being processed seemed unnecessary. However, Adjei-Mensah et al. noted that ViT models are susceptible to low-resolution medical images, suggesting that dataset quality could influence the choice of the mechanism [73].
SQ3: What are the Optimal Learning Conditions for each Model?
The role of pre-training was noteworthy. Among the papers reporting superior or comparable transformer performances, approximately 59% (19 out of 32) described some form of pre-training. In contrast, only 25% (one out of four) of the papers favoring CNNs mentioned pre-training. Furthermore, ImageNet was frequently utilized for pre-training. Despite the belief that transformers benefit considerably from pre-training, this review indicates that pre-training on ImageNet may also be effective for medical imaging models.
SQ4: Which Architecture Demonstrates Greater Robustness?
Regarding the most important SQ, when machine learning models are applied in real-world scenarios, it is unlikely that the learning and testing domains will be identical. A high degree of accuracy is required even when the two domains differ, underscoring the need for robustness. Only three studies used independent external test datasets to assess robustness [56, 67, 83]. Among these, two studies found comparable results, with one favoring the transformer. The similarity in results demonstrates the slight advantage of CNNs in terms of prediction accuracy. Appropriate model selection for each task can result in CNNs outperforming attention-based models in terms of robustness. However, this necessitates the availability of sufficient external test datasets in the selection process.
Central Research Question: When Building a Machine Learning Model Using Medical Images as Input, Which Architecture Should be Used: CNNs or Transformers?
In conclusion, attention-based models are generally recommended when deciding whether to use convolution-based or transformer-based models in machine learning applications for processing medical images. This preference is based on various factors in addition to the above SQs. The first is the superior transparency of the attention-based models, which is critical for user confidence. Attention maps are excellent tools that can provide users with detailed insights. The second is the rapid development of foundation models that employ attention-based mechanisms, as exemplified by Meta’s SAM and MedSAM models [98, 99]. These models are anticipated to be central to future developments. Finally, addressing the question of whether mixed CNN and attention models should be adopted, our advice is against it unless there is a specific reason for adopting them. This recommendation is based on two primary reasons: first, complicating the model could impede the use of pre-trained models, which is critical for achieving high accuracy with attention-based models, and even if a successful model is developed, reusability for others could be challenging. Second, there is no evidence suggesting that mixed models are more robust.
Limitations
This review is subject to certain limitations related to the chosen search terms. Given the relative novelty of attention-based models in the field, the available literature could potentially be skewed toward more favorable findings for these models. The newness of research on attention-based models might lead to an overrepresentation of optimistic results in the literature, as the scientific community tends to swiftly adopt promising new methods. Additionally, there is a potential for subjectivity in the selection of included studies. Despite our efforts to conduct a systematic and unbiased review, the inherent biases and interpretations of researchers may have influenced the results and conclusions drawn from the literature, and these factors cannot be completely eliminated. Referring to similar review articles in the field of general imaging could help mitigate this bias, ensure a more balanced perspective, and strengthen the validity of our conclusions.
Conclusions and Future work
In this systematic review, we have comprehensively examined recent studies in medical image analysis to provide a detailed comparison of CNNs and attention-based models. Our analysis highlights that while both architectures have their distinct strengths, attention-based models hold great promise for advancing the field of medical imaging; however, it is important to recognize that attention-based models are relatively new in the field of medical imaging. This novelty means that their long-term performance and reliability have not yet been fully studied. Therefore, we emphasize the need for further research, particularly longitudinal studies, to determine the consistent effectiveness and potential limitations of attention-based models over time. In addition, future work should focus on refining these models, exploring hybrid architectures that combine the strengths of both CNNs and attention-based models, and evaluating their performance across a broader range of medical imaging tasks and modalities. By addressing these areas, the field can move closer to a more comprehensive understanding of the optimal use of these architectures, ultimately contributing to improved diagnostic accuracy and patient outcomes in clinical practice.
Data Availability
No datasets were generated or analysed during the current study.
References
Alzubaidi L, Zhang J, Humaidi AJ, Al-Dujaili A, Duan Y, Al-Shamma O, Santamaría J, Fadhel MA, Al-Amidie M, Farhan L: Review of deep learning: Concepts, CNN architectures, challenges, applications, future directions. Journal of big Data 2021, 8:1–74. https://doi.org/10.1186/s40537-021-00444-8.
LeCun Y, Bengio Y, Hinton G: Deep learning. Nature 2015, 521(7553):436–444. https://doi.org/10.1038/nature14539.
Bullock J, Cuesta-Lázaro C, Quera-Bofarull A: XNet: a convolutional neural network (CNN) implementation for medical x-ray image segmentation suitable for small datasets. Medical Imaging 2019: Biomedical Applications in Molecular, Structural, and Functional Imaging 2019, 10953:453–463. https://doi.org/10.48550/arXiv.1812.00548.
Dozen A, Komatsu M, Sakai A, Komatsu R, Shozu K, Machino H, Yasutomi S, Arakaki T, Asada K, Kaneko S et al: Image Segmentation of the Ventricular Septum in Fetal Cardiac Ultrasound Videos Based on Deep Learning Using Time-Series Information. Biomolecules 2020, 10(11):1526. https://doi.org/10.3390/biom10111526.
Farooq A, Anwar S, Awais M, Rehman S: A deep CNN based multi-class classification of Alzheimer’s disease using MRI. 2017 IEEE International Conference on Imaging systems and techniques (IST) 2017:1–6. https://doi.org/10.1109/IST.2017.8261460.
Jinnai S, Yamazaki N, Hirano Y, Sugawara Y, Ohe Y, Hamamoto R: The Development of a Skin Cancer Classification System for Pigmented Skin Lesions Using Deep Learning. Biomolecules 2020, 10(8):1123. https://doi.org/10.3390/biom10081123.
Kobayashi K, Hataya R, Kurose Y, Miyake M, Takahashi M, Nakagawa A, Harada T, Hamamoto R: Decomposing Normal and Abnormal Features of Medical Images for Content-Based Image Retrieval of Glioma Imaging. Medical Image Analysis 2021, 74:102227. https://doi.org/10.1016/j.media.2021.102227.
Komatsu M, Sakai A, Komatsu R, Matsuoka R, Yasutomi S, Shozu K, Dozen A, Machino H, Hidaka H, Arakaki T et al: Detection of Cardiac Structural Abnormalities in Fetal Ultrasound Videos Using Deep Learning. Applied Sciences 2021, 11(1):371. https://doi.org/10.3390/app11010371.
Milletari F, Ahmadi S-A, Kroll C, Plate A, Rozanski V, Maiostre J, Levin J, Dietrich O, Ertl-Wagner B, Bötzel K: Hough-CNN: Deep learning for segmentation of deep brain regions in MRI and ultrasound. Computer Vision and Image Understanding 2017, 164:92–102. https://doi.org/10.48550/arXiv.1601.07014.
Yamada M, Saito Y, Imaoka H, Saiko M, Yamada S, Kondo H, Takamaru H, Sakamoto T, Sese J, Kuchiba A et al: Development of a real-time endoscopic image diagnosis support system using deep learning technology in colonoscopy. Sci Rep 2019, 9(1):14465. https://doi.org/10.1038/s41598-019-50567-5.
Yadav D, Rathor S: Bone fracture detection and classification using deep learning approach. 2020 International Conference on Power Electronics & IoT Applications in Renewable Energy and its Control (PARC) 2020:282–285. https://doi.org/10.1109/PARC49193.2020.236611.
Rahman T, Chowdhury ME, Khandakar A, Islam KR, Islam KF, Mahbub ZB, Kadir MA, Kashem S: Transfer learning with deep convolutional neural network (CNN) for pneumonia detection using chest X-ray. Applied Sciences 2020, 10(9):3233. https://doi.org/10.3390/app10093233.
Hamamoto R, Suvarna K, Yamada M, Kobayashi K, Shinkai N, Miyake M, Takahashi M, Jinnai S, Shimoyama R, Sakai A et al: Application of Artificial Intelligence Technology in Oncology: Towards the Establishment of Precision Medicine. Cancers (Basel) 2020, 12(12):3532. https://doi.org/10.3390/cancers12123532.
Asada K, Kobayashi K, Joutard S, Tubaki M, Takahashi S, Takasawa K, Komatsu M, Kaneko S, Sese J, Hamamoto R: Uncovering Prognosis-Related Genes and Pathways by Multi-Omics Analysis in Lung Cancer. Biomolecules 2020, 10(4):524. https://doi.org/10.3390/biom10040524.
Kobayashi K, Bolatkan A, Shiina S, Hamamoto R: Fully-Connected Neural Networks with Reduced Parameterization for Predicting Histological Types of Lung Cancer from Somatic Mutations. Biomolecules 2020, 10(9):1249. https://doi.org/10.3390/biom10091249.
Takahashi S, Asada K, Takasawa K, Shimoyama R, Sakai A, Bolatkan A, Shinkai N, Kobayashi K, Komatsu M, Kaneko S et al: Predicting Deep Learning Based Multi-Omics Parallel Integration Survival Subtypes in Lung Cancer Using Reverse Phase Protein Array Data. Biomolecules 2020, 10(10):1460. https://doi.org/10.3390/biom10101460.
Shin TY, Kim H, Lee J-H, Choi J-S, Min H-S, Cho H, Kim K, Kang G, Kim J, Yoon S: Expert-level segmentation using deep learning for volumetry of polycystic kidney and liver. Investigative and clinical urology 2020, 61(6):555. https://doi.org/10.4111/icu.20200086.
Arab A, Chinda B, Medvedev G, Siu W, Guo H, Gu T, Moreno S, Hamarneh G, Ester M, Song X: A fast and fully-automated deep-learning approach for accurate hemorrhage segmentation and volume quantification in non-contrast whole-head CT. Scientific Reports 2020, 10(1):19389. https://doi.org/10.1038/s41598-020-76459-7
Williams DP: On the use of tiny convolutional neural networks for human-expert-level classification performance in sonar imagery. IEEE Journal of Oceanic Engineering 2020, 46(1):236–260. https://doi.org/10.1109/JOE.2019.2963041.
Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D: Grad-cam: Visual explanations from deep networks via gradient-based localization. Proceedings of the IEEE international conference on computer vision 2017:618–626. https://doi.org/10.48550/arXiv.1610.02391.
Takahashi S, Takahashi M, Kinoshita M, Miyake M, Kawaguchi R, Shinojima N, Mukasa A, Saito K, Nagane M, Otani R et al: Fine-Tuning Approach for Segmentation of Gliomas in Brain Magnetic Resonance Images with a Machine Learning Method to Normalize Image Differences among Facilities. Cancers (Basel) 2021, 13(6). https://doi.org/10.3390/cancers13061415.
Nam H, Lee H, Park J, Yoon W, Yoo D: Reducing domain gap by reducing style bias. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition 2021:8690–8699. https://doi.org/10.48550/arXiv.1910.11645.
Yan W, Wang Y, Gu S, Huang L, Yan F, Xia L, Tao Q: The domain shift problem of medical image segmentation and vendor-adaptation by Unet-GAN. Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, October 13–17, 2019, Proceedings, Part II 22 2019:623–631. https://doi.org/10.48550/arXiv.1910.13681.
Agarwal P, Nachappa M, Gautam CK: Multi-Scale Recurrent Neural Networks for Medical Image Classification. 2024 International Conference on Optimization Computing and Wireless Communication (ICOCWC) 2024:1–6. https://doi.org/10.1109/ICOCWC60930.2024.10470694.
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I: Attention is all you need. Advances in neural information processing systems 2017, 30. https://doi.org/10.48550/arXiv.1706.03762.
Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, Dehghani M, Minderer M, Heigold G, Gelly S: An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:201011929 2020. https://doi.org/10.48550/arXiv.2010.11929.
Liu Y, Wu Y-H, Sun G, Zhang L, Chhatkuli A, Van Gool L: Vision transformers with hierarchical attention. arXiv preprint arXiv:210603180 2021. https://doi.org/10.48550/arXiv.2106.03180.
Han K, Wang Y, Chen H, Chen X, Guo J, Liu Z, Tang Y, Xiao A, Xu C, Xu Y: A survey on vision transformer. IEEE transactions on pattern analysis and machine intelligence 2022, 45(1):87–110. https://doi.org/10.1109/TPAMI.2022.3152247.
Hatamizadeh A, Yin H, Heinrich G, Kautz J, Molchanov P: Global context vision transformers. International Conference on Machine Learning 2023:12633–12646. https://doi.org/10.48550/arXiv.2206.09959.
He K, Gan C, Li Z, Rekik I, Yin Z, Ji W, Gao Y, Wang Q, Zhang J, Shen D: Transformers in medical image analysis. Intelligent Medicine 2023, 3(1):59–78. https://doi.org/10.1016/j.imed.2022.07.002.
Barzekar H, Patel Y, Tong L, Yu Z: MultiNet with Transformers: A Model for Cancer Diagnosis Using Images. arXiv preprint arXiv:230109007 2023. https://doi.org/10.48550/arXiv.2301.09007.
Stassin S, Corduant V, Mahmoudi SA, Siebert X: Explainability and Evaluation of Vision Transformers: An In-Depth Experimental Study. Electronics 2023, 13(1):175. https://doi.org/10.3390/electronics13010175.
Chetoui M, Akhloufi MA: Explainable vision transformers and radiomics for covid-19 detection in chest x-rays. Journal of Clinical Medicine 2022, 11(11):3013. https://doi.org/10.3390/jcm11113013.
Dipto SM, Reza MT, Rahman MNJ, Parvez MZ, Barua PD, Chakraborty S: An XAI Integrated Identification System of White Blood Cell Type Using Variants of Vision Transformer. International Conference on Interactive Collaborative Robotics 2023:303–315. https://doi.org/10.1007/978-3-031-35308-6_26.
Cao Y-H, Yu H, Wu J: Training vision transformers with only 2040 images. European Conference on Computer Vision 2022:220–237. https://doi.org/10.48550/arXiv.2201.10728.
Lee SH, Lee S, Song BC: Vision transformer for small-size datasets. arXiv preprint arXiv:211213492 2021. https://doi.org/10.48550/arXiv.2112.13492.
Liu Y, Sangineto E, Bi W, Sebe N, Lepri B, Nadai M: Efficient training of visual transformers with small datasets. Advances in Neural Information Processing Systems 2021, 34:23818–23830. https://doi.org/10.48550/arXiv.2106.03746.
Habib G, Saleem TJ, Lall B: Knowledge distillation in vision transformers: A critical review. arXiv preprint arXiv:230202108 2023. https://doi.org/10.48550/arXiv.2302.02108.
Youn E, Prabhu S, Chen S: Compressing Vision Transformers for Low-Resource Visual Learning. arXiv preprint arXiv:230902617 2023. https://doi.org/10.48550/arXiv.2309.02617.
Wang X, Zhang LL, Wang Y, Yang M: Towards efficient vision transformer inference: A first study of transformers on mobile devices. Proceedings of the 23rd Annual International Workshop on Mobile Computing Systems and Applications 2022:1–7. https://doi.org/10.1145/3508396.3512869.
Fukushima K: Neocognitron: A self-organizing neural network model for a mechanism of pattern recognition unaffected by shift in position. Biological Cybernetics 1980, 36(4):193–202. https://doi.org/10.1007/BF00344251.
LeCun Y, Bottou L, Bengio Y, Haffner P: Gradient-based learning applied to document recognition. Proceedings of the IEEE 1998, 86(11):2278–2324. https://doi.org/10.1109/5.726791.
Hamamoto R, Komatsu M, Takasawa K, Asada K, Kaneko S: Epigenetics Analysis and Integrated Analysis of Multiomics Data, Including Epigenetic Data, Using Artificial Intelligence in the Era of Precision Medicine. Biomolecules 2020, 10(1):62. https://doi.org/10.3390/biom10010062.
Krizhevsky A, Sutskever I, Hinton GE: ImageNet classification with deep convolutional neural networks. Communications of the ACM 2017, 60(6):84–90. https://doi.org/10.1145/3065386.
Hossin E, Abdelrahim M, Tanasescu A, Yamada M, Kondo H, Yamada S, Hamamoto R, Marugmae A, Saito Y, Bhandari P: Performance of a novel computer-aided diagnosis system in the characterization of colorectal polyps, and its role in meeting Preservation and Incorporation of Valuable Endoscopic Innovations standards set by the American Society of Gastrointestinal Endoscopy. DEN Open 2023, 3(1):e178. https://doi.org/10.1002/deo2.178.
Asada K, Komatsu M, Shimoyama R, Takasawa K, Shinkai N, Sakai A, Bolatkan A, Yamada M, Takahashi S, Machino H et al: Application of Artificial Intelligence in COVID-19 Diagnosis and Therapeutics. Journal of Personalized Medicine 2021, 11(9):886. https://doi.org/10.3390/jpm11090886.
Dabeer S, Khan MM, Islam S: Cancer diagnosis in histopathological image: CNN based approach. Informatics in Medicine Unlocked 2019, 16:100231. https://doi.org/10.1016/j.imu.2019.100231.
Hashimoto N, Fukushima D, Koga R, Takagi Y, Ko K, Kohno K, Nakaguro M, Nakamura S, Hontani H, Takeuchi I: Multi-scale domain-adversarial multiple-instance CNN for cancer subtype classification with unannotated histopathological images. Proceedings of the IEEE/CVF conference on computer vision and pattern recognition 2020:3852–3861. https://doi.org/10.48550/arXiv.2001.01599.
Lin T, Wang Y, Liu X, Qiu X: A survey of transformers. AI open 2022, 3:111–132. https://doi.org/10.1016/j.aiopen.2022.10.001.
Bahdanau D, Cho K, Bengio Y: Neural machine translation by jointly learning to align and translate. arXiv preprint arXiv:14090473 2014. https://doi.org/10.48550/arXiv.1409.0473.
Mondal AK, Bhattacharjee A, Singla P, Prathosh A: xViTCOS: explainable vision transformer based COVID-19 screening using radiography. IEEE Journal of Translational Engineering in Health and Medicine 2021, 10:1–10. https://doi.org/10.1109/JTEHM.2021.3134096
Ikromjanov K, Bhattacharjee S, Hwang Y-B, Sumon RI, Kim H-C, Choi H-K: Whole slide image analysis and detection of prostate cancer using vision transformers. 2022 international conference on artificial intelligence in information and communication (ICAIIC) 2022:399–402. https://doi.org/10.1109/ICAIIC54071.2022.9722635.
Liu Z, Lin Y, Cao Y, Hu H, Wei Y, Zhang Z, Lin S, Guo B: Swin transformer: Hierarchical vision transformer using shifted windows. Proceedings of the IEEE/CVF international conference on computer vision 2021:10012–10022. https://doi.org/10.48550/arXiv.2103.14030.
Snyder H: Literature review as a research methodology: An overview and guidelines. Journal of business research 2019, 104:333–339. https://doi.org/10.1016/j.jbusres.2019.07.039.
Nafisah SI, Muhammad G, Hossain MS, AlQahtani SA: A Comparative Evaluation between Convolutional Neural Networks and Vision Transformers for COVID-19 Detection. Mathematics 2023, 11(6):1489. https://doi.org/10.3390/math11061489.
Deininger L, Stimpel B, Yuce A, Abbasi-Sureshjani S, Schönenberger S, Ocampo P, Korski K, Gaire F: A comparative study between vision transformers and CNNs in digital pathology. arXiv preprint arXiv:220600389 2022. https://doi.org/10.48550/arXiv.2206.00389.
Wu Y, Qi S, Sun Y, Xia S, Yao Y, Qian W: A vision transformer for emphysema classification using CT images. Physics in Medicine & Biology 2021, 66(24):245016. https://doi.org/10.1088/1361-6560/ac3dc8
Xing X, Liang G, Zhang Y, Khanal S, Lin A-L, Jacobs N: Advit: Vision transformer on multi-modality pet images for alzheimer disease diagnosis. 2022 IEEE 19th International Symposium on Biomedical Imaging (ISBI) 2022:1–4. https://doi.org/10.1109/ISBI52829.2022.9761584.
Xin C, Liu Z, Zhao K, Miao L, Ma Y, Zhu X, Zhou Q, Wang S, Li L, Yang F et al: An improved transformer network for skin cancer classification. Comput Biol Med 2022, 149:105939. https://doi.org/10.1016/j.compbiomed.2022.105939.
Usman M, Zia T, Tariq A: Analyzing transfer learning of vision transformers for interpreting chest radiography. Journal of digital imaging 2022, 35(6):1445–1462. https://doi.org/10.1007/s10278-022-00666-z
Carcagnì P, Leo M, Del Coco M, Distante C, De Salve A: Convolution Neural Networks and Self-Attention Learners for Alzheimer Dementia Diagnosis from Brain MRI. Sensors 2023, 23(3):1694. https://doi.org/10.3390/s23031694.
Ambita AAE, Boquio ENV, Naval Jr PC: Covit-gan: vision transformer forcovid-19 detection in ct scan imageswith self-attention gan forDataAugmentation. International Conference on Artificial Neural Networks 2021:587–598. https://doi.org/10.1155/2022/8925930.
Xiao J, Bai Y, Yuille A, Zhou Z: Delving into masked autoencoders for multi-label thorax disease classification. Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision 2023:3588–3600. https://doi.org/10.48550/arXiv.2210.12843.
Tyagi K, Pathak G, Nijhawan R, Mittal A: Detecting Pneumonia using Vision Transformer and comparing with other techniques. 2021 5th International Conference on Electronics, Communication and Aerospace Technology (ICECA) 2021:12–16. https://doi.org/10.1109/ICECA52323.2021.9676146.
Rajaraman S, Zamzmi G, Folio LR, Antani S: Detecting tuberculosis-consistent findings in lateral chest X-rays using an ensemble of CNNs and vision transformers. Frontiers in Genetics 2022, 13:864724. https://doi.org/10.3389/fgene.2022.864724
Kumar NS, Karthikeyan BR: Diabetic Retinopathy Detection using CNN, Transformer and MLP based Architectures. 2021 International Symposium on Intelligent Signal Processing and Communication Systems (ISPACS) 2021:1–2. https://doi.org/10.1109/ISPACS51563.2021.9651024.
Playout C, Duval R, Boucher MC, Cheriet F: Focused attention in transformers for interpretable classification of retinal images. Medical Image Analysis 2022, 82:102608. https://doi.org/10.1016/j.media.2022.102608
Okolo GI, Katsigiannis S, Ramzan N: IEViT: An enhanced vision transformer architecture for chest X-ray image classification. Computer Methods and Programs in Biomedicine 2022, 226:107141. https://doi.org/10.1016/j.cmpb.2022.107141.
Kermany D, Zhang K, Goldbaum M: Labeled optical coherence tomography (oct) and chest x-ray images for classification. Mendeley data 2018, 2(2):651.
Feng H, Yang B, Wang J, Liu M, Yin L, Zheng W, Yin Z, Liu C: Identifying malignant breast ultrasound images using ViT-patch. Applied Sciences 2023, 13(6):3489. https://doi.org/10.3390/app13063489.
Al-Dhabyani W, Gomaa M, Khaled H, Fahmy A: Dataset of breast ultrasound images. Data in brief 2020, 28:104863. https://doi.org/10.1016/j.dib.2019.104863
Cho P, Dash S, Tsaris A, Yoon H-J: Image transformers for classifying acute lymphoblastic leukemia. Medical Imaging 2022: Computer-Aided Diagnosis 2022, 12033:633–639. https://doi.org/10.1117/12.2611496.
Adjei-Mensah I, Zhang X, Baffour AA, Agyemang IO, Yussif SB, Agbley BLY, Sey C: Investigating vision transformer models for low-resolution medical image recognition. 2021 18th International Computer Conference on Wavelet Active Media Technology and Information Processing (ICCWAMTIP) 2021:179–183. https://doi.org/10.1109/ICCWAMTIP53232.2021.9674065.
Jiang Z, Dong Z, Wang L, Jiang W: Method for diagnosis of acute lymphoblastic leukemia based on ViT-CNN ensemble model. Computational Intelligence and Neuroscience 2021, 2021. https://doi.org/10.1155/2021/7529893
Pachetti E, Colantonio S, Pascali MA: On the effectiveness of 3D vision transformers for the prediction of prostate cancer aggressiveness. International Conference on Image Analysis and Processing 2022:317–328. https://doi.org/10.1007/978-3-031-13324-4_27.
Matsoukas C, Haslum JF, Söderberg M, Smith K: Pretrained ViTs Yield Versatile Representations For Medical Images. arXiv preprint arXiv:230307034 2023. https://doi.org/10.48550/arXiv.2303.07034.
Aitazaz T, Tubaishat A, Al-Obeidat F, Shah B, Zia T, Tariq A: Transfer learning for histopathology images: an empirical study. Neural Computing and Applications 2023, 35(11):7963–7974. https://doi.org/10.1007/s00521-022-07516-7.
Mohan NJ, Murugan R, Goel T, Roy P: ViT-DR: Vision Transformers in Diabetic Retinopathy Grading Using Fundus Images. 2022 IEEE 10th Region 10 Humanitarian Technology Conference (R10-HTC) 2022:167–172. https://doi.org/10.1109/R10-HTC54060.2022.9930027.
Wang H, Ji Y, Song K, Sun M, Lv P, Zhang T: ViT-P: Classification of genitourinary syndrome of menopause from OCT images based on vision transformer models. IEEE Transactions on Instrumentation and Measurement 2021, 70:1–14. https://doi.org/10.1109/TIM.2021.3122121.
Wu J, Hu R, Xiao Z, Chen J, Liu J: Vision Transformer-based recognition of diabetic retinopathy grade. Medical Physics 2021, 48(12):7850–7863. https://doi.org/10.1002/mp.15312
Tanzi L, Audisio A, Cirrincione G, Aprato A, Vezzetti E: Vision transformer for femur fracture classification. Injury 2022, 53(7):2625–2634. https://doi.org/10.48550/arXiv.2108.03414.
Gheflati B, Rivaz H: Vision transformers for classification of breast ultrasound images. Annu Int Conf IEEE Eng Med Biol Soc 2022:480–483. https://doi.org/10.1109/EMBC48229.2022.9871809.
Murphy ZR, Venkatesh K, Sulam J, Yi PH: Visual Transformers and Convolutional Neural Networks for Disease Classification on Radiographs: A Comparison of Performance, Sample Efficiency, and Hidden Stratification. Radiology: Artificial Intelligence 2022, 4(6):e220012. https://doi.org/10.1148/ryai.220012
Liu W, Li C, Rahaman MM, Jiang T, Sun H, Wu X, Hu W, Chen H, Sun C, Yao Y: Is the aspect ratio of cells important in deep learning? A robust comparison of deep learning methods for multi-scale cytopathology cell image classification: From convolutional neural networks to visual transformers. Computers in biology and medicine 2022, 141:105026. https://doi.org/10.1016/j.compbiomed.2021.105026.
Mok TC, Chung A: Affine medical image registration with coarse-to-fine vision transformer. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition 2022:20835–20844. https://doi.org/10.48550/arXiv.2203.15216.
Karimi D, Vasylechko SD, Gholipour A: Convolution-free medical image segmentation using transformers. Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part I 24 2021:78–88. https://doi.org/10.1007/978-3-030-87193-2_8.
Nguyen C, Asad Z, Deng R, Huo Y: Evaluating transformer-based semantic segmentation networks for pathological image segmentation. Medical Imaging 2022: Image Processing 2022, 12032:942–947. https://doi.org/10.1117/12.2611177.
Karimi D, Dou H, Gholipour A: Medical image segmentation using transformer networks. IEEE Access 2022, 10:29322–29332. https://doi.org/10.1109/ACCESS.2022.3156894.
Gulzar Y, Khan SA: Skin lesion segmentation based on vision transformers and convolutional neural networks—A comparative study. Applied Sciences 2022, 12(12):5990. https://doi.org/10.3390/app12125990.
Hatamizadeh A, Nath V, Tang Y, Yang D, Roth HR, Xu D: Swin unetr: Swin transformers for semantic segmentation of brain tumors in mri images. International MICCAI Brainlesion Workshop 2021:272–284. https://doi.org/10.48550/arXiv.2201.01266.
Cao H, Wang Y, Chen J, Jiang D, Zhang X, Tian Q, Wang M: Swin-unet: Unet-like pure transformer for medical image segmentation. European conference on computer vision 2022:205–218. https://doi.org/10.48550/arXiv.2105.05537.
Hagos MT, Kant S: Transfer learning based detection of diabetic retinopathy from small dataset. arXiv preprint arXiv:190507203 2019. https://doi.org/10.48550/arXiv.1905.07203.
Ronneberger O, Fischer P, Brox T: U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the International Conference on Medical image computing and computer-assisted intervention 2015:1505.04597. https://doi.org/10.1007/978-3-319-24574-4_28.
happyharrycn M, Phil Culliton, Poonam Yadav, Sangjune Laurence Lee: UW-Madison GI Tract Image Segmentation. Kaggle. https://kaggle.com/competitions/uw-madison-gi-tract-image-segmentation 2022.
Baid U, Ghodasara S, Mohan S, Bilello M, Calabrese E, Colak E, Farahani K, Kalpathy-Cramer J, Kitamura FC, Pati S: The rsna-asnr-miccai brats 2021 benchmark on brain tumor segmentation and radiogenomic classification. arXiv preprint arXiv:210702314 2021. https://doi.org/10.48550/arXiv.2107.02314.
Codella N, Rotemberg V, Tschandl P, Celebi ME, Dusza S, Gutman D, Helba B, Kalloo A, Liopyris K, Marchetti M: Skin lesion analysis toward melanoma detection 2018: A challenge hosted by the international skin imaging collaboration (isic). arXiv preprint arXiv:190203368 2019. https://doi.org/10.48550/arXiv.1902.03368.
Tang G, Müller M, Rios A, Sennrich R: Why self-attention? a targeted evaluation of neural machine translation architectures. arXiv preprint arXiv:180808946 2018. https://doi.org/10.48550/arXiv.1808.08946.
Kirillov A, Mintun E, Ravi N, Mao HZ, Rolland C, Gustafson L, Xiao TT, Whitehead S, Berg AC, Lo WY et al: Segment Anything. Ieee I Conf Comp Vis 2023:3992–4003. https://doi.org/10.1109/Iccv51070.2023.00371.
Ma J, He Y, Li F, Han L, You C, Wang B: Segment anything in medical images. Nature Communications 2024, 15(1):654. https://doi.org/10.1038/s41467-024-44824-z
Acknowledgements
We thank all the members of R. Hamamoto’s laboratory for providing valuable advice and a comfortable environment.
Funding
This work was supported by BRIDGE (programs for bridging the gap between R&D and the ideal society (Society 5.0) and generating economic and social value), JSPS Kakenhi (22K16700), and MEXT subsidy for the Advanced Integrated Intelligence Platform.
Author information
Authors and Affiliations
Contributions
ST, KT, and RH made substantial contributions to the study conception and design; ST, YS, and NK conducted the searches and the screening process, and assessed the quality appraisal of the included studies. MS and RH supervised the entire study. ST, KT, KI, YK, RA, NT, AB, NS, HM, KK, MK, SK, MS, RH drafted the manuscript. All authors critically revised the manuscript, approved the final version to be published, and agree to be accountable for aspects of the work.
Corresponding author
Ethics declarations
Ethics Approval
This systematic review is based on the analysis of previously published data. No primary data collection of human subject involvement was performed, ensuring compliance with ethical guidelines and regulations. Therefore, ethical approval was not required.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Takahashi, S., Sakaguchi, Y., Kouno, N. et al. Comparison of Vision Transformers and Convolutional Neural Networks in Medical Image Analysis: A Systematic Review. J Med Syst 48, 84 (2024). https://doi.org/10.1007/s10916-024-02105-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10916-024-02105-8