Abstract
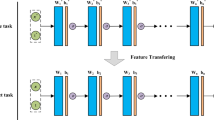
Transfer learning for partial differential equations (PDEs) is to develop a pre-trained neural network that can be used to solve a wide class of PDEs. Existing transfer learning approaches require much information about the target PDEs such as its formulation and/or data of its solution for pre-training. In this work, we propose to design transferable neural feature spaces for the shallow neural networks from purely function approximation perspectives without using PDE information. The construction of the feature space involves the re-parameterization of the hidden neurons and uses auxiliary functions to tune the resulting feature space. Theoretical analysis shows the high quality of the produced feature space, i.e., uniformly distributed neurons. We use the proposed feature space as the pre-determined feature space of a random feature model, and use existing least squares solvers to obtain the weights of the output layer. Extensive numerical experiments verify the outstanding performance of our method, including significantly improved transferability, e.g., using the same feature space for various PDEs with different domains and boundary conditions, and the superior accuracy, e.g., several orders of magnitude smaller mean squared error than the state of the art methods.








Similar content being viewed by others
Data availability
Enquiries about data availability should be directed to the authors.
Notes
Note that the dimension of the feature space is the sum of both space and time dimensions since it doesn’t differ them.
BFGS can alleviate ill-conditioning by exploiting the second-order information, e.g., the approximate Hessian.
References
Raissi, M., Perdikaris, P., Karniadakis, G.E.: Physics-informed neural networks: a deep learning framework for solving forward and inverse problems involving nonlinear partial differential equations. J. Comput. Phys. 378, 686–707 (2019)
Weinan, E., Bing, Y.: The deep Ritz method: a deep learning-based numerical algorithm for solving variational problems. Commun. Math. Stat. 6(1), 1–12 (2018)
Long, Z., Lu, Y., Ma, X., Dong, B.: PDE-Net: Learning PDEs from data. In: International Conference on Machine Learning, pp. 3214–3222, (2018)
Zang, Y., Bao, G., Ye, X., Zhou, H.: Weak adversarial networks for high dimensional partial differential equations. J. Comput. Phys. 411, 109409 (2020)
Li, Z., Kovachki, N.B., Azizzadenesheli, K., Bhattacharya, K., Stuart, A., Anandkumar, A. et al.: Fourier neural operator for parametric partial differential equations. In: International Conference on Learning Representations, (2021)
Li, Z., Kovachki, N., Azizzadenesheli, K., Liu, B., Stuart, A., Bhattacharya, K., Anandkumar, A.: Multipole graph neural operator for parametric partial differential equations. Adv. Neural. Inf. Process. Syst. 33, 6755–6766 (2020)
Lu, L., Jin, P., Pang, G., Zhang, Z., Karniadakis, G.E.: Learning nonlinear operators via DeepONet based on the universal approximation theorem of operators. Nat. Mach. Intell. 3(3), 218–229 (2021)
Gin, C.R., Shea, D.E., Brunton, S.L., Nathan Kutz, J.: Deepgreen: deep learning of green’s functions for nonlinear boundary value problems. Sci. Rep. 11(1), 1–14 (2021)
Zhang, X., Cheng, T., Ju, L.: Implicit form neural network for learning scalar hyperbolic conservation laws. In: Mathematical and Scientific Machine Learning Conference, pp. 1082–1098, (2021)
Teng, Y., Zhang, X., Wang, Z., Ju, L.: Learning green’s functions of linear reaction-diffusion equations with application to fast numerical solver. In: Mathematical and Scientific Machine Learning Conference, (2022)
Di, L., Patricio, C., Lu, L., Meneveau, C., Karniadakis, G.E., Zaki, T.A.: Neural operator prediction of linear instability waves in high-speed boundary layers. J. Comput. Phys. 474, 111793 (2023)
Souvik Lal Chakraborty: Transfer learning based multi-fidelity physics informed deep neural network. J. Comput. Phys. 426, 109942 (2020)
Desai, S., Mattheakis, M., Joy, H., Protopapas, P., Roberts, S.J.: One-shot transfer learning of physics-informed neural networks. arXiv:2110.11286, (2021)
Cybenko, G.: Approximation by superpositions of a sigmoidal function. Math. Control Signals Syst. 2(4), 303–314 (1989)
Lagaris, I.E., Likas, A.C., Papageorgiou, D.G.: Neural-network methods for boundary value problems with irregular boundaries. IEEE Trans. Neural Netw. 11(5), 1041–1049 (2000)
Pakdaman, M., Ahmadian, A., Effati, S., Salahshour, S., Baleanu, D.: Solving differential equations of fractional order using an optimization technique based on training artificial neural network. Appl. Math. Comput. 293, 81–95 (2017)
Piscopo, M.L., Spannowsky, M., Waite, P.: Solving differential equations with neural networks: applications to the calculation of cosmological phase transitions. Phys. Rev. D 100(1), 016002 (2019)
Sun, Y., Gilbert, A.C., Tewari, A.: On the approximation capabilities of relu neural networks and random relu features. arxiv:1810.04374 (2018)
Liu, Yuxuan, McCalla, S.G., Schaeffer, H.: Random feature models for learning interacting dynamical systems, (2022)
Chen, J., Chi, X., Weinan, E., Zhouwang, Y.: The random feature method, Bridging traditional and machine learning-based algorithms for solving pdes (2022)
Dissanayake, M., Phan-Thien, N.: Neural-network-based approximations for solving partial differential equations. Commun. Numer. Methods Eng. 10(3), 195–201 (1994)
Lagaris, I.E., Likas, A., Fotiadis, D.I.: Artificial neural networks for solving ordinary and partial differential equations. IEEE Trans. Neural Netw. 9(5), 987–1000 (1998)
Lu, L., Meng, X., Mao, Z., Karniadakis, G.E.: Deepxde: A deep learning library for solving differential equations. SIAM Rev. 63(1), 208–228 (2021)
Anitescu, C., Atroshchenko, E., Alajlan, N., Rabczuk, T.: Artificial neural network methods for the solution of second order boundary value problems. Comput. Mater. Continua 59(1), 345–359 (2019)
Zhao, J., Wright, C.L.: Solving allen-cahn and cahn-hilliard equations using the adaptive physics informed neural networks. Commun. Comput. Phys. 29, 930–954 (2021)
Krishnapriyan, A., Gholami, A., Zhe, S., Kirby, R., Mahoney, M.W.: Characterizing possible failure modes in physics-informed neural networks. Adv. Neural Inf. Process. Syst. 34, 26548–60 (2021)
Sirignano, J., Spiliopoulos, K.: DGM: a deep learning algorithm for solving partial differential equations. J. Comput. Phys. 375, 1339–1354 (2018)
Long, Z., Lu, Y., Dong, B.: PDE-Net 2.0: Learning PDEs from data with a numeric-symbolic hybrid deep network. J. Comput. Phys. 399, 108925 (2019)
Chen, T., Chen, H.: Universal approximation to nonlinear operators by neural networks with arbitrary activation functions and its application to dynamical systems. IEEE Trans. Neural Netw. 6(4), 911–917 (1995)
Wang, S., Wang, H., Perdikaris, P.: Learning the solution operator of parametric partial differential equations with physics-informed deeponets. Sci. Adv. 7(40), eabi8605 (2021)
Li, Z., Zheng, H., Kovachki, N., Jin, D., Chen, H., Liu, B., Azizzadenesheli, K., Anandkumar, A.: Physics-informed neural operator for learning partial differential equations. arXiv preprint arXiv:2111.03794, (2021)
Jin, P., Meng, S., Lu, L.: Mionet: learning multiple-input operators via tensor product. SIAM J. Sci. Comput. 44(6), A3490–A3514 (2022)
Nelsen, N.H., Stuart, A.M.: The random feature model for input-output maps between banach spaces. SIAM J. Sci. Comput. 43(5), A3212–A3243 (2021)
Liu, F., Huang, X., Chen, Y., Suykens, J.A.K.: Random features for kernel approximation: a survey on algorithms, theory, and beyond. IEEE Trans. Pattern Anal. Mach. Intell. 44(10), 7128–7148 (2022)
Bach, F.: On the equivalence between kernel quadrature rules and random feature expansions. J. Mach. Learn. Res. 18(1), 714–751 (2017)
Karniadakis, G.E., Kevrekidis, I.G., Lu, L., Perdikaris, P., Wang, S., Yang, L.: Physics-informed machine learning. Nat. Rev. Phys. 3(6), 422–440 (2021)
McDonald, T., Álvarez, M.: Compositional modeling of nonlinear dynamical systems with ode-based random features. In: M. Ranzato, A. Beygelzimer, Y. Dauphin, P.S. Liang, and J. Wortman Vaughan, editors, Advances in Neural Information Processing Systems, volume 34, pp. 13809–13819. Curran Associates, Inc., (2021)
Arora, R., Basu, A., Mianjy, P., Mukherjee, A.: Understanding deep neural networks with rectified linear units. arXiv preprint arXiv:1611.01491, (2016)
Daubechies, I., DeVore, R., Foucart, S., Hanin, B., Petrova, G.: Nonlinear approximation and (deep) relu networks. Constr. Approx. 55(1), 127–172 (2022)
Pascanu, R., Montufar, G., Bengio, Y.: On the number of response regions of deep feed forward networks with piece-wise linear activations. arXiv preprint arXiv:1312.6098, (2013)
Montufar, G.F., Pascanu, R., Cho, K., Bengio, Y.: On the number of linear regions of deep neural networks. Adv. Neural Inf. Process. Syst. 27, 2924–2932 (2014)
Serra, T., Tjandraatmadja, C., Ramalingam, S.: Bounding and counting linear regions of deep neural networks. In: International Conference on Machine Learning, pp. 4558–4566. PMLR, (2018)
Serra, T., Ramalingam, S.: Empirical bounds on linear regions of deep rectifier networks. Procee. AAAI Conf. Artif. Intell. 34, 5628–5635 (2020)
Hanin, B., Rolnick, D.: Complexity of linear regions in deep networks. In: International Conference on Machine Learning, pp. 2596–2604. PMLR, (2019)
Fang, K.W.: Symmetric multivariate and related distributions. CRC Press, Florida (2018)
Acknowledgements
This work is supported by the U.S. Department of Energy, Office of Science, Office of Advanced Scientific Computing Research, Applied Mathematics program, under the contract ERKJ387, and accomplished at Oak Ridge National Laboratory (ORNL), and under the grants DE-SC0022254 and DE-SC0022297. ORNL is operated by UT-Battelle, LLC., for the U.S. Department of Energy under the contract DE-AC05-00OR22725.
Funding
The authors have not disclosed any funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have not disclosed any competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This manuscript has been authored by UT-Battelle, LLC, under contract DE-AC05-00OR22725 with the US Department of Energy (DOE). The US government retains and the publisher, by accepting the article for publication, acknowledges that the US government retains a nonexclusive, paid-up, irrevocable, worldwide license to publish or reproduce the published form of this manuscript, or allow others to do so, for US government purposes. DOE will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan.
Appendices
Appendix
Definitions of the PDEs in Sect. 3.2
The definitions of the PDEs considered in Sect. 3.2 are given below.
The Poisson’s equation considered in case \((C_1)\)–\((C_5)\) is defined by
where the exact solution for the 2D settings, i.e., \((C_1)\)–\((C_4)\), is \(u(\varvec{x}) = \sin (2\pi x_1)\sin (2\pi x_2)\sin (2\pi x_3)\), and the exact solution for the 3D setting, i.e., \((C_5)\), is \(u(\varvec{x}) = \sin (2\pi x_1)\sin (2\pi x_2)\). The forcing term \(f(\varvec{x})\) can be obtained by applying the Laplacian operator to the exact solution. The domains of computation for \((C_1)\)–\((C_5)\) are given below:
- (\(C_1\)):
-
A 2D rectangular domain: \(\varOmega = [-1,1]^2\);
- (\(C_2\)):
-
A 2D circular domain: \(\varOmega = B_1(\varvec{0})\);
- (\(C_3\)):
-
A 2D L-shaped domain: \(\varOmega = [-1,1]^2 \backslash [0,1]^2\);
- (\(C_4\)):
-
A 2D annulus domain: \(\varOmega = B_1(\varvec{0}) \backslash B_{0.5}(\varvec{0})\);
- (\(C_5\)):
-
A 3D box domain \(\varOmega = [-1,1]^3\).
We consider the Dirichlet boundary condition in the experiments, where the boundary condition \(g(\varvec{x})\) in Eq. (1) can be obtained by restricting the exact solution on the boundary of \(\varOmega \). Figure 7 illustrates how to place the domains of computation into the unit ball for the test cases \((C_1)\) – \((C_4)\) to use the transferable feature space.
Illustration of how to place the domains of computation for the test cases \((C_1)\) – \((C_4)\) in Sect. 3.2 into the unit ball to use the transferable feature space to solve the Poisson’s equation in different domains
The steady-state Navier–Stokes equation considered in case \((C_6)\) is defined by:
where \(\varvec{u} = (v_1, v_2)\) represents the velocity, p is the pressure, \(\nu \) is the viscosity and \(Re = 1/\nu \) is the Reynold’s number. The domain of computation is \(\varOmega = [-0.5,1]\times [-0.5,1.5]\) with Direchilet boundary condition. We consider the Kovasznay flow problem that has the exact solution, i.e.,
where \(\lambda = \frac{1}{2\nu } - \sqrt{\frac{1}{4\nu ^2} + 4\pi ^2}\) and the Reynold’s number is set to 40. The Dirichlet boundary condition can be obtained by restricting the exact solution on the boundary of \(\varOmega \).
The Fokker-Planck equation considered in case \((C_7)\) and \((C_8)\) is defined by
where the coefficients \(b(t,\varvec{x})\), \(\sigma \), \(g(\varvec{x})\) and the exact solutions are
-
\((C_7)\): \(b(x,t) = 2 \cos {(3 t)}\), \(\sigma =0.3\), \(u(x,0) = p(x; 0, 0.4^2)\) and \(u(x,t) = p(x; \frac{2\sin { (3t)}}{3}, 0.4^2 + t 0.3^2)\), where \(p(x; \mu , \varSigma )\) denote the Gaussian density with mean \(\mu \) and variance \(\varSigma \).
-
\((C_8)\): \(b(x_1, x_2, t) = [\sin (2\pi t), \cos (2\pi t)]^T\), \(\sigma =0.3\), \(u(x_1, x_2, 0) = p(x; [0,0], 0.4^2 \textbf{I}_2)\), and \(u(x_1, x_2, t) = p(x; [-\frac{\cos (2\pi t)-1}{2\pi }, \frac{\sin (2\pi t)}{2\pi } ], (0.4^2 + t 0.3^2)\textbf{I}_2)\), where \(p(x; \mu , \varSigma )\) is the Gaussian density with mean \(\mu \) and variance \(\varSigma \).
The wave equation considered in case \((C_9)\) is defined by
where \(c = 1/(16\pi ^2)\). The domain of computation is \(\varOmega = [0,1] \times [0,2]\); the exact solution is
Setup of the Experiments in Sect. 3.2
We specify the setup for the test cases \((C_1)\) to \((C_9)\) as follows:
-
\((C_1)\): We evaluate the loss function in Eq. (19) on a \(50 \times 50\) uniform mesh in \(\varOmega = [-1,1]^2\), i.e., \(J_1 = 2500\) in Eq. (19), and on 200 uniformly distributed points on \(\partial \varOmega \), i.e., \(J_2 = 200\). After solving the least squares problem, we compute the error, i.e., the results shown in Fig. 5 on a test set of 10,000 uniformly distributed random locations in \(\varOmega \).
-
\((C_2)\): We evaluate the loss function in Eq. (19) on a \(50 \times 50\) uniform mesh in \(\varOmega = [-1,1]^2\) and mask off the grid points outside the domain \(\varOmega = B_1(\varvec{0})\), i.e., \(J_1 = 1876\), and evaluate the boundary loss on 200 uniformly distributed points on \(\partial \varOmega \), i.e., \(J_2 = 200\). After solving the least squares problem, we compute the error, i.e., the results shown in Fig. 5 on a test set of 10,000 uniformly distributed random locations in \(\varOmega \).
-
\((C_3)\): We evaluate the loss function in Eq. (19) on a \(50 \times 50\) uniform mesh in \(\varOmega = [-1,1]^2\) and mask off the grid points outside the domain \(\varOmega = [-1,1]^2 \backslash [0,1]^2\), i.e., \(J_1 = 1875\), and evaluate the boundary loss on 200 uniformly distributed points on \(\partial \varOmega \), i.e., \(J_2 = 200\). After solving the least squares problem, we compute the error, i.e., the results shown in Fig. 5 on a test set of 10,000 uniformly distributed random locations in \(\varOmega \).
-
\((C_4)\): We evaluate the loss function in Eq. (19) on a \(50 \times 50\) uniform mesh in \(\varOmega = [-1,1]^2\) and mask off the grid points outside the domain \(\varOmega = B_1(\varvec{0}) \backslash B_{0.5}(\varvec{0})\), i.e., \(J_1 = 1408\), and evaluate the boundary loss on 200 uniformly distributed points on \(\partial \varOmega \), i.e., \(J_2 = 200\). After solving the least squares problem, we compute the error, i.e., the results shown in Fig. 5 on a test set of 10,000 uniformly distributed random locations in \(\varOmega \).
-
\((C_5)\): We evaluate the loss function in Eq. (19) on a 10,000 uniformly distributed random locations in \(\varOmega = [-1,1]^3\), i.e., \(J_1 = 10000\), and evaluate the boundary loss on 2400 uniformly distributed points on \(\partial \varOmega \), i.e., \(J_2 = 2400\), 400 points on each side of \(\varOmega \). After solving the least squares problem, we compute the error, i.e., the results shown in Fig. 5 on a test set of 10,000 uniformly distributed random locations in \(\varOmega \).
-
\((C_6)\): We evaluate the loss function in Eq. (19) on a \(50 \times 50\) uniform mesh in \(\varOmega = [-0.5,1]\times [-0.5,1.5]\), i.e., \(J_1 = 2500\) in Eq. (19), and on 200 uniformly distributed points on \(\partial \varOmega \) (50 points on each side of the box), i.e., \(J_2 = 200\). We use Pichard iteration to handle the nonlinearity. Specifically, the residual loss is defined by
$$\begin{aligned} loss = {\varvec{u}_\text {NN}^{k-1}} \cdot \nabla {\varvec{u}_\text {NN}^{k}} + \nabla p_\text {NN}^k - \nu \varDelta {\varvec{u}_\text {NN}^{k}}, \end{aligned}$$where k is the Picard iteration number. In the k-th iteration, the nonlinear term \({\varvec{u}_\text {NN}^{k-1}} \cdot \nabla {\varvec{u}_\text {NN}^{k}}\) becomes linear due to the use of \({\varvec{u}_\text {NN}^{k-1}}\). After solving the least squares problem, we compute the error, i.e., the results shown in Fig. 5 on a test set of 10,000 uniformly distributed random locations in \(\varOmega \).
-
\((C_7)\): The domain of computation is \((t,x) \in [0,1]\times [-2,2]\). We evaluate the loss function on a 50 (time) \(\times \) 200 (space) = 10,000 grid points in the domain \(\varOmega \). We use the absorbing boundary condition in the spatial domain. We have a total of 3000 samples on the boundary of \(\varOmega \), i.e., 1000 samples for each of u(x, 0), u(2, t) and \(u(-2,t)\). After solving the least squares problem, we compute the error, i.e., the results shown in Fig. 5 on a test set of 10,000 uniformly distributed random locations in \(\varOmega \).
-
\((C_8)\): The domain of computation is \(t \in [0,1]\) and \((x_1,x_2) \in [-2,2]^2\). We evaluate the loss function on 10,000 uniformly selected random points in the domain \(\varOmega \). We use the absorbing boundary condition in the spatial domain. In terms of samples on the boundary, we have \(50 \times 50 = 2500\) grid points for the initial condition \(u(x_1, x_2, 0)\), \(20(\text {time}) \times 50(\text {space}) = 1000\) grid points for each of \(u(\pm 2, x_2, t)\) and \(u(x_1,\pm 2, t)\). After solving the least squares problem, we compute the error, i.e., the results shown in Fig. 5 on a test set of 10,000 uniformly distributed random locations in \(\varOmega \).
-
\((C_9)\): We evaluate the loss function in Eq. (19) on \(50\text {(time)} \times 100\text {(space)} = 2500\) grid points in domain, i.e., \(J_1 = 10,000\), and evaluate the boundary loss on 1000 uniformly distributed points on \(\partial \varOmega \), i.e., \(J_2 = 1500\), 500 points on each side of \(\varOmega \). After solving the least squares problem, we compute the error, i.e., the results shown in Fig. 5 on a test set of 10,000 uniformly distributed random locations in \(\varOmega \).
We use the standard least squares solver torch.linalg.lstsq in Pytorch to solve all the least squares problems. Our code is implemented using Pytorch on a workstation with an NVIDIA Tesla V100 GPU.
Setup for PINN. The code for PINN is included in the supplementary material. For each test case, PINN uses exactly the same setting as TransNet, including network architecture, loss function, and data, to ensure a fair comparison. In terms of training, we set the learning rate to 0.001 with a decrease factor of 0.7 every 1000 epochs. We first use Adam optimizer to train the neural networks for 5000 epochs, which gives us the results in Fig. 5 labeled by “PINN:Adam”. Then we continue training the network using LBFGS for another 200 iterations, which gives us the results in Fig. 5 labeled by “PINN:Adam+BFGS”.
Setup for the random feature models. The random feature model uses exactly the same setting as TransNet, including network architecture, loss function, and data, to ensure a fair comparison. The parameters \(\{\varvec{w}_m, b_m\}_{m=1}^M\) are determined by the default initialization methods in Pytorch, and the parameters in the output layer are obtained by the least squares solver torch.linalg.lstsq in Pytorch.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, Z., Bao, F., Ju, L. et al. Transferable Neural Networks for Partial Differential Equations. J Sci Comput 99, 2 (2024). https://doi.org/10.1007/s10915-024-02463-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10915-024-02463-y