Abstract
Solving high-dimensional Bayesian inverse problems (BIPs) with the variational inference (VI) method is promising but still challenging. The main difficulties arise from two aspects. First, VI methods approximate the posterior distribution using a simple and analytic variational distribution, which makes it difficult to estimate complex spatially-varying parameters in practice. Second, VI methods typically rely on gradient-based optimization, which can be computationally expensive or intractable when applied to BIPs involving partial differential equations (PDEs). To address these challenges, we propose a novel approximation method for estimating the high-dimensional posterior distribution. This approach leverages a deep generative model to learn a prior model capable of generating spatially-varying parameters. This enables posterior approximation over the latent variable instead of the complex parameters, thus improving estimation accuracy. Moreover, to accelerate gradient computation, we employ a differentiable physics-constrained surrogate model to replace the adjoint method. The proposed method can be fully implemented in an automatic differentiation manner. Numerical examples demonstrate two types of log-permeability estimation for flow in heterogeneous media. The results show the validity, accuracy, and high efficiency of the proposed method.


















Similar content being viewed by others
Data Availibility
The datasets generated during and/or analyzed during the current study are available in the github repository, https://github.com/xiayzh/VI-DGP.
References
Barajas-Solano, D.A., Tartakovsky, A.M.: Approximate bayesian model inversion for PDEs with heterogeneous and state-dependent coefficients. J. Comput. Phys. 395, 247–262 (2019)
Bilionis, I., Zabaras, N., Konomi, B.A., Lin, G.: Multi-output separable gaussian process: towards an efficient, fully bayesian paradigm for uncertainty quantification. J. Comput. Phys. 241, 212–239 (2013)
Blei, D.M., Kucukelbir, A., McAuliffe, J.D.: Variational inference: a review for statisticians. J. Am. Stat. Assoc. 112(518), 859–877 (2017)
Bora, A., Jalal, A., Price, E., Dimakis, A.G.: Compressed sensing using generative models. In: International Conference on Machine Learning, pp. 537–546. PMLR (2017)
Bui-Thanh, T., Girolami, M.: Solving large-scale PDE-constrained bayesian inverse problems with Riemann manifold Hamiltonian monte Carlo. Invers. Probl. 30(11), 114014 (2014)
Chen, P., Ghattas, O.: Stein variational reduced basis bayesian inversion. SIAM J. Sci. Comput. 43(2), A1163–A1193 (2021)
Cotter, S.L., Roberts, G.O., Stuart, A.M., White, D.: Mcmc methods for functions: modifying old algorithms to make them faster. Stat. Sci. 28(3), 424–446 (2013)
Cui, T., Marzouk, Y.M., Willcox, K.E.: Data-driven model reduction for the bayesian solution of inverse problems. Int. J. Numer. Meth. Eng. 102(5), 966–990 (2015)
Driggs, D., Liang, J., Schönlieb, C.B.: On biased stochastic gradient estimation. J. Mach. Learn. Res. 23(1), 1057–1099 (2022)
Engl, H.W., Hanke, M., Neubauer, A.: Regularization of Inverse Problems, vol. 375. Springer Science & Business Media (1996)
Fan, Y., Ying, L.: Solving inverse wave scattering with deep learning. arXiv preprint arXiv:1911.13202 (2019)
Geneva, N., Zabaras, N.: Modeling the dynamics of PDE systems with physics-constrained deep auto-regressive networks. J. Comput. Phys. 403, 109056 (2020)
Goodfellow, I., Pouget-Abadie, J., Mirza, M., Xu, B., Warde-Farley, D., Ozair, S., Courville, A., Bengio, Y.: Generative adversarial nets. Adv. Neural Inf. Process. Syst. 27 (2014)
Guha, N., Wu, X., Efendiev, Y., Jin, B., Mallick, B.K.: A variational bayesian approach for inverse problems with skew-t error distributions. J. Comput. Phys. 301, 377–393 (2015)
Hairer, M., Stuart, A.M., Vollmer, S.J.: Spectral gaps for a metropolis–hastings algorithm in infinite dimensions. Ann. Appl. Probab. 24(6), 2455–2490 (2014)
Heusel, M., Ramsauer, H., Unterthiner, T., Nessler, B., Hochreiter, S.: Gans trained by a two time-scale update rule converge to a local nash equilibrium. Adv. Neural Inf. Process. Syst. 30 (2017)
Jalal, A., Arvinte, M., Daras, G., Price, E., Dimakis, A.G., Tamir, J.: Robust compressed sensing MRI with deep generative priors. Adv. Neural Inf. Process. Syst. 34, 14938–14954 (2021)
Jia, J., Zhao, Q., Xu, Z., Meng, D., Leung, Y.: Variational bayes’ method for functions with applications to some inverse problems. SIAM J. Sci. Comput. 43(1), A355–A383 (2021)
Kaipio, J., Somersalo, E.: Statistical and Computational Inverse Problems, vol. 160. Springer Science & Business Media (2006)
Khoo, Y., Ying, L.: Switchnet: a neural network model for forward and inverse scattering problems. SIAM J. Sci. Comput. 41(5), A3182–A3201 (2019)
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014)
Kingma, D.P., Welling, M.: Auto-encoding variational bayes. arXiv preprint arXiv:1312.6114 (2013)
Laloy, E., Hérault, R., Jacques, D., Linde, N.: Training-image based geostatistical inversion using a spatial generative adversarial neural network. Water Resour. Res. 54(1), 381–406 (2018)
Laloy, E., Hérault, R., Lee, J., Jacques, D., Linde, N.: Inversion using a new low-dimensional representation of complex binary geological media based on a deep neural network. Adv. Water Resour. 110, 387–405 (2017)
Li, S., Xia, Y., Liu, Y., Liao, Q.: A deep domain decomposition method based on Fourier features. J. Comput. Appl. Math. 423, 114963 (2023)
Liao, Q., Li, J.: An adaptive reduced basis anova method for high-dimensional bayesian inverse problems. J. Comput. Phys. 396, 364–380 (2019)
Lu, L., Jin, P., Pang, G., Zhang, Z., Karniadakis, G.E.: Learning nonlinear operators via deeponet based on the universal approximation theorem of operators. Nat. Mach. Intell. 3(3), 218–229 (2021)
Lye, K.O., Mishra, S., Ray, D., Chandrashekar, P.: Iterative surrogate model optimization (ISMO): An active learning algorithm for PDE constrained optimization with deep neural networks. Comput. Meth. Appl. Mech. Eng. 374, 113575 (2021)
Martin, J., Wilcox, L.C., Burstedde, C., Ghattas, O.: A stochastic newton MCMC method for large-scale statistical inverse problems with application to seismic inversion. SIAM J. Sci. Comput. 34(3), A1460–A1487 (2012)
Marzouk, Y.M., Najm, H.N., Rahn, L.A.: Stochastic spectral methods for efficient bayesian solution of inverse problems. J. Comput. Phys. 224(2), 560–586 (2007)
Metropolis, N., Rosenbluth, A.W., Rosenbluth, M.N., Teller, A.H., Teller, E.: Equation of state calculations by fast computing machines. J. Chem. Phys. 21(6), 1087–1092 (1953)
Mo, S., Zabaras, N., Shi, X., Wu, J.: Deep autoregressive neural networks for high-dimensional inverse problems in groundwater contaminant source identification. Water Resour. Res. 55(5), 3856–3881 (2019)
Mo, S., Zabaras, N., Shi, X., Wu, J.: Integration of adversarial autoencoders with residual dense convolutional networks for estimation of non-gaussian hydraulic conductivities. Water Resourc. Res. 56(2), e2019WR026082 (2020)
Mo, S., Zhu, Y., Zabaras, N., Shi, X., Wu, J.: Deep convolutional encoder–decoder networks for uncertainty quantification of dynamic multiphase flow in heterogeneous media. Water Resourc. Res. 55(1), 703–728 (2019)
Padmanabha, G.A., Zabaras, N.: Solving inverse problems using conditional invertible neural networks. J. Comput. Phys. 433, 110194 (2021)
Patel, D.V., Ray, D., Oberai, A.A.: Solution of physics-based bayesian inverse problems with deep generative priors. Comput. Meth. Appl. Mech. Eng. 400, 115428 (2022)
Povala, J., Kazlauskaite, I., Febrianto, E., Cirak, F., Girolami, M.: Variational bayesian approximation of inverse problems using sparse precision matrices. Comput. Meth. Appl. Mech. Eng. 393, 114712 (2022)
Raissi, M., Perdikaris, P., Karniadakis, G.E.: Physics-informed neural networks: a deep learning framework for solving forward and inverse problems involving nonlinear partial differential equations. J. Comput. Phys. 378, 686–707 (2019)
Ranganath, R., Gerrish, S., Blei, D.: Black box variational inference. In: Artificial Intelligence and Statistics, pp. 814–822. PMLR (2014)
Rezende, D., Mohamed, S.: Variational inference with normalizing flows. In: International Conference on Machine Learning, pp. 1530–1538. PMLR (2015)
Robert, C.P., Casella, G., Casella, G.: Monte Carlo Statistical Methods, vol. 2. Springer (1999)
Roeder, G., Wu, Y., Duvenaud, D.K.: Sticking the landing: Simple, lower-variance gradient estimators for variational inference. Adv. Neural Inf. Process. Syst. 30 (2017)
Stuart, A.M.: Inverse problems: a bayesian perspective. Acta Numer. 19, 451–559 (2010)
Sun, L., Gao, H., Pan, S., Wang, J.X.: Surrogate modeling for fluid flows based on physics-constrained deep learning without simulation data. Comput. Meth. Appl. Mech. Eng. 361, 112732 (2020)
Tarantola, A.: Inverse Problem Theory and Methods for Model Parameter Estimation, vol. 89. SIAM (2005)
Tripathy, R.K., Bilionis, I.: Deep UQ: Learning deep neural network surrogate models for high dimensional uncertainty quantification. J. Comput. Phys. 375, 565–588 (2018)
Tsilifis, P., Bilionis, I., Katsounaros, I., Zabaras, N.: Computationally efficient variational approximations for bayesian inverse problems. J. Verif. Valid. Uncertain. Quantif. 1(3), 031004 (2016)
Wan, J., Zabaras, N.: A bayesian approach to multiscale inverse problems using the sequential monte Carlo method. Invers. Probl. 27(10), 105004 (2011)
Wang, K., Bui-Thanh, T., Ghattas, O.: A randomized maximum a posteriori method for posterior sampling of high dimensional nonlinear bayesian inverse problems. SIAM J. Sci. Comput. 40(1), A142–A171 (2018)
Wang, L., Chan, Y.C., Ahmed, F., Liu, Z., Zhu, P., Chen, W.: Deep generative modeling for mechanistic-based learning and design of metamaterial systems. Comput. Meth. Appl. Mech. Eng. 372, 113377 (2020)
Wang, S., Bhouri, M.A., Perdikaris, P.: Fast pde-constrained optimization via self-supervised operator learning. arXiv preprint arXiv:2110.13297 (2021)
Warner, J.E., Aquino, W., Grigoriu, M.D.: Stochastic reduced order models for inverse problems under uncertainty. Comput. Meth. Appl. Mech. Eng. 285, 488–514 (2015)
Xia, Y., Zabaras, N.: Bayesian multiscale deep generative model for the solution of high-dimensional inverse problems. J. Comput. Phys. 455, 111008 (2022)
Xiu, D., Karniadakis, G.E.: Modeling uncertainty in flow simulations via generalized polynomial chaos. J. Comput. Phys. 187(1), 137–167 (2003)
Xu, Z., Xia, Y., Liao, Q.: A domain-decomposed vae method for bayesian inverse problems. arXiv preprint arXiv:2301.05708 (2023)
Yan, L., Zhou, T.: Stein variational gradient descent with local approximations. Comput. Meth. Appl. Mech. Eng. 386, 114087 (2021)
Yang, K., Guha, N., Efendiev, Y., Mallick, B.K.: Bayesian and variational bayesian approaches for flows in heterogeneous random media. J. Comput. Phys. 345, 275–293 (2017)
Zhang, C., Bütepage, J., Kjellström, H., Mandt, S.: Advances in variational inference. IEEE Trans. Patt. Anal. Mach. Intell. 41(8), 2008–2026 (2018)
Zhdanov, M.S.: Geophysical Inverse Theory and Regularization Problems, vol. 36. Elsevier (2002)
Zhu, Y., Zabaras, N.: Bayesian deep convolutional encoder-decoder networks for surrogate modeling and uncertainty quantification. J. Comput. Phys. 366, 415–447 (2018)
Zhu, Y., Zabaras, N., Koutsourelakis, P.S., Perdikaris, P.: Physics-constrained deep learning for high-dimensional surrogate modeling and uncertainty quantification without labeled data. J. Comput. Phys. 394, 56–81 (2019)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This work is supported by the National Natural Science Foundation of China (No. 12071291), the Science and Technology Commission of Shanghai Municipality (No. 20JC1414300), the Natural Science Foundation of Shanghai (No. 20ZR1436200) and A*STAR AME Programmatic Fund: Explainable Physics-based AI for Engineering Modelling & Design (Grant No. A20H5b0142).
Appendices
Appendix A: The Derivation of VAE and the Reparameterization Trick
Note that Maximizing \({\mathcal {L}}({\varvec{\theta },\varvec{\phi }};\varvec{k})\) in Eq. (8) will maximize the marginal log-likelihood and also make approximation \(q_{\varvec{\phi }}(\varvec{z}|\varvec{k})\) close to the true posterior \(p_{\varvec{\theta }}(\varvec{z}|\varvec{k})\). So we can maximize \({\mathcal {L}}({\varvec{\theta },\varvec{\phi }};\varvec{k})\) rather than the marginal log-likelihood for computational convenience [22]. For the given training dataset \({\textbf{K}}\), we can write the ELBO for any given \(\varvec{k}^{(i)}\) as
Obviously, these two terms play different roles in optimization. The first term is the expected log-likelihood \(\log p_{\varvec{\theta }}(\varvec{k}|\varvec{z})\), where \(\varvec{z}\) is sampled from the probabilistic encoder \(q_{\varvec{\phi }}(\varvec{z}|\varvec{k})\). Maximizing this term enforces \(p_{\varvec{\theta }}(\varvec{k}|\varvec{z})\) to assign most of the probability density close to the original \(\varvec{k}\). The second term aims to minimize the KL divergence between \(q_{\varvec{\phi }}(\varvec{z}|\varvec{k})\) and \(p_{\varvec{\theta }}(\varvec{z})\), which regularizes the probabilistic encoder \(q_{\varvec{\phi }}(\varvec{z}|\varvec{k}^{(i)})\) to resemble the prior distribution \(p_{\varvec{\theta }}(\varvec{z})\). These two terms are the reconstruction term and the regularization term, respectively. For loss computation and parameters \(\varvec{\phi }\) and \(\varvec{\theta }\) optimization, we still need to specify the distributions for \(p_{\varvec{\theta }}(\varvec{k}|\varvec{z})\), \(q_{\varvec{\phi }}(\varvec{z}|\varvec{k})\), and \(p_{\varvec{\theta }}(\varvec{z})\) in Eq. (25). Typically, one can assign a simple isotropic Gaussian distribution as the prior distribution, e.g.,
Ideally, an appropriate probabilistic encoder \(q_{\varvec{\phi }}(\varvec{z}|\varvec{k})\) should be able to approximate the target distribution \(p_{\varvec{\theta }}(\varvec{z})\) well. Additionally, a Gaussian distribution with a diagonal covariance can be selected as the variational distribution:
where \(\varvec{\mu }_{\phi }(\varvec{k})\) and \(\varvec{\sigma }_{\phi }(\varvec{k})\) are computed by the encoder neural networks. The KL divergence term in Eq. (25) has an analytic form [22] since both \(p_{\varvec{\theta }}(\varvec{z})\) and \(q_{\varvec{\phi }}(\varvec{z}|\varvec{k})\) are the factorized Gaussian distribution. The distribution \(p_{\varvec{\theta }}(\varvec{k}|\varvec{z})\) usually depends on the training data. In this paper, we select the Gaussian distribution \({\mathcal {N}}\left( \varvec{k}; {\mathcal {G}}_{\varvec{\theta }}(\varvec{z}), {\varvec{I}}\right) \) for the probabilistic decoder, where \({\mathcal {G}}_{\varvec{\theta }}(\varvec{z})\) is the output of the decoder neural networks. The stochastic gradient-based method is applied for large-scale training data to realize the joint optimization for \(\{\varvec{\theta }, \varvec{\phi }\}\) using the objective function \({\mathcal {L}}({\varvec{\theta },\varvec{\phi }};\varvec{k})\). The reconstruction term in Eq. (25) involves the expectation computation, which is tackled by Monte Carlo estimation. The gradient \(\nabla _{\varvec{\theta }}{\mathbb {E}}_{q_{\varvec{\phi }}(\varvec{z}|\varvec{k})} [\log p_{\varvec{\theta }}(\varvec{k}|\varvec{z})]\) can be estimated directly, where the latent variable \(\varvec{z}\) is randomly sampled from \(q_{\varvec{\phi }}(\varvec{z}|\varvec{k})\) for expectation approximation. However, the gradient \(\nabla _{\varvec{\phi }}{\mathbb {E}}_{q_{\varvec{\phi }}(\varvec{z}|\varvec{k})} [\log p_{\varvec{\theta }}(\varvec{k}|\varvec{z})]\) is difficult to obtain. One cannot swap the gradient and the expectation since the expectation with respect to the distribution \(q_{\varvec{\phi }}(\varvec{z}|\varvec{k})\) is a function of \(\varvec{\phi }\). The score function estimator [3, 39] can be applied for gradient estimation, but its high variance leads to a slow optimization process. An alternative differentiable estimator with low variance is the reparameterization trick [22, 40], where the latent variable \(\varvec{z}\) is represented by a deterministic transformation \(\varvec{z}= \varvec{g}_{\varvec{\phi }} (\varvec{\epsilon };\varvec{k})\).
The differentiable transformation \(\varvec{g}_{\varvec{\phi }} (\varvec{\epsilon };\varvec{k})\) maps the auxiliary random noise to the Gaussian distribution in Eq. (27) by the following procedure:
where \(\odot \) denotes the element-wise product and \(\pi (\varvec{\epsilon }) = {\mathcal {N}}({\textbf{0}}, {\textbf{I}})\). Then the random variable \(\varvec{z}\) only depends on two deterministic outputs of the encoder neural networks by introducing an auxiliary random variable \(\varvec{\epsilon }\). Since the operators \(+\) and \(\odot \) are differentiable, the gradient \(\nabla _{\varvec{\phi }}{\mathbb {E}}_{q_{\varvec{\phi }}(\varvec{z}|\varvec{k})} [\log p_{\varvec{\theta }}(\varvec{k}|\varvec{z})]\) is available. It can be written as
which can be directly estimated by the Monte Carlo method with L samples drawn from \(\pi (\varvec{\epsilon })\). Then the ELBO in Eq. (25) can be rewritten as
where \(\varvec{z}^{(i,l)}\) is the \(l-\)th sample drawn from \(q_{\varvec{\phi }}(\varvec{z}|\varvec{k}^{(i)})\). To improve computational efficiency, the training of neural networks usually adopts the minibatch stochastic gradient-based method, where the training dataset is divided into many subsets. Each subset contains n data points for each iteration. The optimization objective function in each iteration can be written as
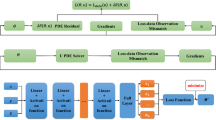
One can apply the stochastic gradient-based method, such as Adam [21], to optimize the probabilistic encoder \(q_{\varvec{\phi }}(\varvec{z}|\varvec{k})\) and the probabilistic decoder \(p_{\varvec{\theta }}(\varvec{k}|\varvec{z})\) using the above objective function. Figure 16 depicts a schematic illustration of the VAE model and the reparameterization trick.
The schematic illustration of the VAE model and the reparameterization trick. The spatially-varying parameter \(\varvec{k}\) is mapped to a latent variable \(\varvec{z}\) by the probabilistic encoder \(q_{\varvec{\phi }}(\varvec{z}|\varvec{k})\). In turn, the latent variable \(\varvec{z}\) is mapped to the parameter \(\varvec{k}\) by the probabilistic decoder \(p_{\varvec{\theta }}(\varvec{k}|\varvec{z})\)
Appendix B: The Network Architectures for the Encoder and Decoder in VAE
In this work, we use fully-connected neural networks as the encoder and decoder for both Gaussian and channel cases. Table 3 illustrates the implemented neural networks for the encoder and decoder. For the decoder, we use ReLU and Sigmoid as the activation function for the Gaussian and channel cases, respectively. Additionally, for the channel case, we apply an extra Sigmoid activation function for the last layer of the decoder model, which ensures that the output values are within the interval [0, 1]. h denotes the number of neurons in the encoder’s hidden layer, which will also define the dimensionality of the latent variable \(\varvec{z}\). We set h to 256 and 512 for the Gaussian and channel cases, respectively.
Appendix C: The Network Architectures for the Physics-Constrained Surrogate Model
We can rewrite the loss function in discretization form for the given PDEs in Eq. (21) and Eq. (22). The PDEs loss and boundary loss in Eq. (19) can be written as
respectively, where \(n_b\) boundary samples include \(n_{bl}\) samples of left boundary \({\mathcal {D}}_l\), \(n_{br}\) samples of right boundary \({\mathcal {D}}_r\), \(n_{bt}\) samples of top boundary \({\mathcal {D}}_t\), and \(n_{bb}\) samples of bottom boundary \({\mathcal {D}}_b\).
The network architectures applied in this paper are based on previous works [60, 61]. These works perform greatly in uncertainty quantification tasks for the flow in heterogeneous media. The main architectures are shown in Table 4. The number of dense layers in the three dense blocks is 6, 8, 6, with a growth rate of 16. Each dense layer contains a Conv block (Batch-ReLU-Conv). Encoding 1, Decoding 1, and Decoding 2 have 2, 2, 3 Conv blocks, respectively. The nearest mode is used for the upsampling operator in the decoding layers.
Appendix D: The pCN Algorithm for MCMC Simulation
We employ the pCN algorithm to explore the posterior distribution, which is the reference method for the proposed approach. The details are shown in the Algorithm 4, where the forward model \({\mathcal {F}}(\cdot )\) can be either the learned neural network surrogate or the finite element method. These correspond to MCMC-NN and MCMC-FEM in the experiments, respectively.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xia, Y., Liao, Q. & Li, J. VI-DGP: A Variational Inference Method with Deep Generative Prior for Solving High-Dimensional Inverse Problems. J Sci Comput 97, 16 (2023). https://doi.org/10.1007/s10915-023-02328-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10915-023-02328-w
Keywords
- Inverse problems
- Variational inference
- Deep generative model
- Physics-constrained surrogate
- Gradient approximation