Abstract
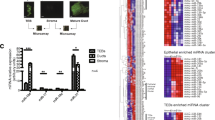
Mammary stem/progenitor cells are fundamental for mammary gland development and function. However, much remains to be elucidated regarding their function in mammals beyond the traditionally studied rodents, human, and to a lesser extent, ruminants. Due to the growing appreciation for microRNAs (miRNAs) as regulators of stem cells and their progenitors, we compared miRNA expression in mammary stem/progenitor cells from mammals with varying mammary stem/progenitor activity in vitro, in order to identify miRNA candidates that regulate stem/progenitor self-renewal and function. Mammosphere-derived epithelial cells (MDECs), which are primary cell lines enriched in mammary stem and progenitor cells, were generated from six mammalian species (i.e., cow, human, pig, horse, dog, and rat) and small RNA sequencing was performed. We identified 9 miRNAs that were significantly differentially expressed in MDEC cultures with a low versus high mammary stem/progenitor activity. miR-92b-3p was selected for functional follow-up studies, as this miRNA is understudied in primary mammary cells but has well-described gene targets that are known to regulate mammary stem/progenitor activity. Altering the expression of miR-92b-3p in MDECs from species with low stem/progenitor activity (human and cow) and those with high stem/progenitor activity (dog and rat) via inhibition and overexpression, respectively, resulted in significantly decreased mammosphere formation of human MDECs, but showed no significant effects in cow, dog, or rat MDECs. This study is the first to perform small RNA sequencing in MDECs from various mammals and highlights that conserved miRNAs can have different functions in mammary stem/progenitor cells across species.







Similar content being viewed by others
Data Availability
The miRNA sequencing data are accessible through GEOSeries # GSE126424 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE126424).
References
Visvader JE, Stingl J. Mammary stem cells and the differentiation hierarchy: Current status and perspectives. Genes Dev. 2014;28:1143–58. https://doi.org/10.1101/gad.242511.114.
Rauner G, Ledet MM, Van de Walle GR. Conserved and variable: Understanding mammary stem cells across species. Cytom Part A. 2018;93A:125–36. https://doi.org/10.1002/cyto.a.23190.
Spaas JH, Chiers K, Bussche L, Burvenich C, Van De Walle GR. Stem/Progenitor Cells in Non-Lactating Versus Lactating Equine Mammary Gland. Stem Cells Dev. 2012;21:3055–67. https://doi.org/10.1089/scd.2012.0042.
Bussche L, Rauner G, Antonyak M, Syracuse B, McDowell M, Brown AMC, et al. Microvesicle-mediated wnt/β-catenin signaling promotes interspecies mammary stem/progenitor cell growth. J Biol Chem. 2016;291:24390–405. https://doi.org/10.1074/jbc.m116.726117.
O’Brien J, Hayder H, Zayed Y, Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol. 2018;9. https://doi.org/10.3389/fendo.2018.00402.
Ha M, Pang M, Agarwal V, Chen ZJ. Interspecies regulation of MicroRNAs and their targets. Biochim Biophys Acta. 2008;1779:735–42. https://doi.org/10.1016/j.bbagrm.2008.03.004.
Li N, Long B, Han W, Yuan S, Wang K. MicroRNAs. Important regulators of stem cells. Stem Cell Res Ther. 2017;8. https://doi.org/10.1186/s13287-017-0551-0.
Berezikov E. Evolution of microRNA diversity and regulation in animals. Nat Rev Genet. 2011;12:846–60. https://doi.org/10.1038/nrg3079.
Shehata M, Teschendorff A, Sharp G, Novcic N, Russell IA, Avril S, et al. Phenotypic and functional characterisation of the luminal cell hierarchy of the mammary gland. Breast Cancer Res. 2012;14:R134. https://doi.org/10.1186/bcr3334.
Wu A, Dong Q, Gao H, Shi Y, Chen Y, Zhang F, et al. Characterization of mammary epithelial stem/progenitor cells and their changes with aging in common marmosets. Sci Rep. 2016;6:32190. https://doi.org/10.1038/srep32190.
Lombardo Y, de Giorgio A, Coombes CR, Stebbing J, Castellano L. Mammosphere formation assay from human breast cancer tissues and cell lines. J Vis Exp. 2015;e52671. https://doi.org/10.3791/52671.
Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–70. https://doi.org/10.1101/gad.1061803.
Rauner G, Barash I. Cell hierarchy and lineage commitment in the bovine mammary gland. PLoS ONE. 2012;7:e30113. https://doi.org/10.1371/journal.pone.0030113.
Ledet MM, Oswald M, Anderson R, Van de Walle GR. Differential signaling pathway activation in 7,12-dimethylbenz[a] anthracene (DMBA)-treated mammary stem/progenitor cells from species with varying mammary cancer incidence. Oncotarget. 2018;9:32761–74. https://doi.org/10.18632/oncotarget.25988.
Hu Y, Smyth GK. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–8. https://doi.org/10.1016/j.jim.2009.06.008.
Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–7. https://doi.org/10.1038/nature04496.
Meier-Abt F, Milani E, Roloff T, Brinkhaus H, Duss S, Meyer DS, et al. Parity induces differentiation and reduces Wnt/Notch signaling ratio and proliferation potential of basal stem/progenitor cells isolated from mouse mammary epithelium. Breast Cancer Res. 2013;15:R36. https://doi.org/10.1186%2Fbcr3419.
Mazzoni E, Adam A, De Kier Joffe EB, Aguirre-Ghiso JA. Immortalized mammary epithelial cells overexpressing protein kinase C γ acquire a malignant phenotype and become tumorigenic in vivo. Mol Cancer Res. 2003;1:776–87.
Bao L, Cardiff RD, Steinbach P, Messer KS, Ellies LG. Multipotent luminal mammary cancer stem cells model tumor heterogeneity. Breast Cancer Res. 2015;17:137. https://doi.org/10.1186/s13058-015-0615-y.
Kanke M, Baran-Gale J, Vallanueva J, Sethupathy P. miRquant 2.0: an Expanded Tool for Accurate Annotation and Quantification of MicroRNAs and their isomiRs from Small RNA-Sequencing Data. J Integr Bioinform. 2016;13:307. https://doi.org/10.2390/biecoll-jib-2016-307.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. https://doi.org/10.1186/s13059-014-0550-8.
Beaver CM, Ahmed A, Masters JR. Clonogenicity: Holoclones and Meroclones Contain Stem Cells. PLoS ONE. 2014;9:e89834. https://doi.org/10.1371/journal.pone.0089834.
Barrandon Y, Green H. Three clonal types of keratinocyte with different capacities for multiplication. Proc Natl Acad Sci U S A. 1987;84:2302–6. https://doi.org/10.1073/pnas.84.8.2302.
Qian N-S, Liu W-H, Lv W-P, Xiang X, Su M, Raut V, et al. Upregulated MicroRNA-92b Regulates the Differentiation and Proliferation of EpCAM-Positive Fetal Liver Cells by Targeting C/EBPß. PLoS ONE. 2013;8:e68004. https://doi.org/10.1371/journal.pone.0068004.
Wu ZB, Cai L, Lin SJ, Lu JL, Yao Y, Zhou LF. The miR-92b functions as a potential oncogene by targeting on Smad3 in glioblastomas. Brain Res. 2013;1529:16–25. https://doi.org/10.1016/j.brainres.2013.07.031.
Liu F, Sang M, Meng L, Gu L, Liu S, Li J, et al. miR–92b promotes autophagy and suppresses viability and invasion in breast cancer by targeting EZH2. Int J Oncol. 2018;53:1505–15. https://doi.org/10.3892/ijo.2018.4486.
Chen L, Zhuo HZ, Wu JY, Lin LY, Huang ZL, Lu JX, et al. MiR-92b inhibits proliferation and invasion of lung cancer by targeting EZH2. Eur Rev Med Pharmacol Sci. 2020;24:3166–73. https://doi.org/10.26355/eurrev_202003_20683.
Chen Z, Li Z, Jiang C, Jiang X, Zhang J. MiR-92b-3p promotes neurite growth and functional recovery via the PTEN/AKT pathway in acute spinal cord injury. J Cell Physiol. 2019;234:23043–52. https://doi.org/10.1002/jcp.28864.
Ibarra I, Erlich Y, Muthuswamy SK, Sachidanandam R, Hannon GJ. A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev. 2007;21:3243. https://doi.org/10.1101/gad.1616307.
Tian L, Li Y, Wang C, Li Q. Let-7 g-5p regulates mouse mammary cells differentiation and function by targeting PRKCA. J Cell Physiol. 2019;234:10101–10. https://doi.org/10.1002/jcp.27676.
Xuan R, Chao T, Wang A, Zhang F, Sun P, Liu S, et al. Characterization of microRNA profiles in the mammary gland tissue of dairy goats at the late lactation, dry period and late gestation stages. PLoS ONE. 2020;15:e0234427. https://doi.org/10.1371/journal.pone.0234427.
Greene SB, Gunaratne PH, Hammond SM, Rosen JM. A putative role for microRNA-205 in mammary epithelial cell progenitors. J Cell Sci. 2010;123:606–18. https://doi.org/10.1242/jcs.056812.
Lu Y, Cao J, Napoli M, Xia Z, Zhao N, Creighton CJ, et al. miR-205 Regulates Basal Cell Identity and Stem Cell Regenerative Potential During Mammary Reconstitution. Stem Cells. 2018;36:1875–89. https://doi.org/10.1002/stem.2914.
Chao C-H, Chang C-C, Wu M-J, Ko H-W, Wang D, Hung M-C, et al. MicroRNA-205 signaling regulates mammary stem cell fate and tumorigenesis. J Clin Invest. 2014;124:3093–106. https://doi.org/10.1172/jci73351.
Lamarca HL, Visbal AP, Creighton CJ, Liu H, Zhang Y, Behbod F, et al. C/EBPβ Regulates Stem Cell Activity and Specifies Luminal Cell Fate in the Mammary Gland. Stem Cells. 2010;28:535–44. https://doi.org/10.1002/stem.297.
Spike AJ, Rosen JM. C/EBPß Isoform Specific Gene Regulation: It’s a Lot more Complicated than you Think! J Mammary Gland Biol Neoplasia. 2020;25:1–12. https://doi.org/10.1007/s10911-020-09444-5.
Gonzalez ME, Moore HM, Li X, Toy KA, Huang W, Sabel MS, et al. EZH2 expands breast stem cells through activation of NOTCH1 signaling. Proc Natl Acad Sci U S A. 2014;111:3098–103. https://doi.org/10.1073/pnas.1308953111.
Pennington MR, Van de Walle GR. Electric Cell-Substrate Impedance Sensing To Monitor Viral Growth and Study Cellular Responses to Infection with Alphaherpesviruses in Real Time. mSphere. 2017;2:e00039-17. https://doi.org/10.1128/mSphere.00039-17.
Stolwijk JA, Matrougui K, Renken CW, Trebak M. Impedance analysis of GPCR-mediated changes in endothelial barrier function: overview and fundamental considerations for stable and reproducible measurements. Pflügers Arch. 2015;467:2193–218. https://doi.org/10.1007/s00424-014-1674-0.
Szulcek R, Bogaard HJ, van Nieuw Amerongen GP. Electric Cell-substrate Impedance Sensing for the Quantification of Endothelial Proliferation, Barrier Function, and Motility. J Vis Exp. 2014;51300. https://doi.org/10.3791/51300.
Bussche L, Harman RM, Syracuse BA, Plante EL, Lu YC, Curtis TM, et al. Microencapsulated equine mesenchymal stromal cells promote cutaneous wound healing in vitro. Stem Cell Res Ther. 2015;6:66. https://doi.org/10.1186/s13287-015-0037-x.
Harman RM, Patel RS, Fan JC, Park JE, Rosenberg BR, Van de Walle GR. Single-cell RNA sequencing of equine mesenchymal stromal cells from primary donor-matched tissue sources reveals functional heterogeneity in immune modulation and cell motility. Stem Cell Res Ther. 2020;11:524. https://doi.org/10.1186/s13287-020-02043-5.
Harman RM, Curtis TM, Argyle DJ, Coonrod SA, Van de Walle GR. A Comparative Study on the In Vitro Effects of the DNA Methyltransferase Inhibitor 5-Azacytidine (5-AzaC) in Breast/Mammary Cancer of Different Mammalian Species. J Mammary Gland Biol Neoplasia. 2016;21:51–66. https://doi.org/10.1007/s10911-016-9350-y.
Cui Y, Fan Y, Zhao G, Zhang Q, Bao Y, Cui Y, et al. Novel lncRNA PSMG3-AS1 functions as a miR-143-3p sponge to increase the proliferation and migration of breast cancer cells. Oncol Rep. 2020;43:229–39. https://doi.org/10.3892/or.2019.7390.
Zhang ML, Cao MW, Kong LH, Liu J, Wang YH, Song CC, et al. MiR-204-5p promotes lipid synthesis in mammary epithelial cells by targeting SIRT1. Biochem Biophys Res Commun. 2020;533:1490–6. https://doi.org/10.1016/j.bbrc.2020.10.056.
Mor E, Shomron N. Species-specific microRNA regulation influences phenotypic variability: perspectives on species-specific microRNA regulation. BioEssays. 2013;35:881–8. https://doi.org/10.1002/bies.201200157.
Utikal J, Abba M, Novak D, Moniuszko M, Allgayer H. Function and significance of MicroRNAs in benign and malignant human stem cells. Semin Cancer Biol. 2015;35:200–11. https://doi.org/10.1016/j.semcancer.2015.07.001.
Liu S, Patel SH, Ginestier C, Ibarra I, Martin-Trevino R, Bai S, et al. MicroRNA93 regulates proliferation and differentiation of normal and malignant breast stem cells. PLoS Genet. 2012;8:e1002751. https://doi.org/10.1371/journal.pgen.1002751.
Lv C, Li F, Li X, Tian Y, Zhang Y, Sheng X, et al. MiR-31 promotes mammary stem cell expansion and breast tumorigenesis by suppressing Wnt signaling antagonists. Nat Commun. 2017;8:1036. https://doi.org/10.1038/s41467-017-01059-5.
Martignani E, Cravero D, Miretti S, Accornero P, Baratta M. Clonogenic assay allows for selection of a primitive mammary epithelial cell population in bovine. Exp Cell Res. 2015;338:245–50. https://doi.org/10.1016/j.yexcr.2015.08.016.
Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9:201–9. https://doi.org/10.1038/ncb1530.
Janda E, Nevolo M, Lehmann K, Downward J, Beug H, Grieco M. Raf plus TGFβ-dependent EMT is initiated by endocytosis and lysosomal degradation of E-cadherin. Oncogene. 2006;25:7117–30. https://doi.org/10.1038/sj.onc.1209701.
Kim H-J, Litzenburger BC, Cui X, Delgado DA, Grabiner BC, Lin X, et al. Constitutively Active Type I Insulin-Like Growth Factor Receptor Causes Transformation and Xenograft Growth of Immortalized Mammary Epithelial Cells and Is Accompanied by an Epithelial-to-Mesenchymal Transition Mediated by NF-κB and Snail. Mol Cell Biol. 2007;27:3165–75. https://doi.org/10.1128/MCB.01315-06.
Thiery JP, Acloque H, Huang RYJ, Nieto MA. Epithelial-Mesenchymal Transitions in Development and Disease. Cell. 2009;139:871–90. https://doi.org/10.1016/j.cell.2009.11.007.
Bartov E, Jerdan JA, Glaser BM. A simple technique for isolating pure cell populations from mixed primary cultures. J tissue Cult methods. 1988;11:181–3. https://doi.org/10.1007/BF01407311.
Wang H, Agarwal P, Jiang B, Stewart S, Liu X, Liang Y, et al. Bioinspired One Cell Culture Isolates Highly Tumorigenic and Metastatic Cancer Stem Cells Capable of Multilineage Differentiation. Adv Sci. 2020;7:2000259. https://doi.org/10.1002%2Fadvs.202000259.
Yang X, Lin X, Zhong X, Kaur S, Li N, Liang S, et al. Double-Negative Feedback Loop between Reprogramming Factor LIN28 and microRNA let-7 Regulates Aldehyde Dehydrogenase 1–Positive Cancer Stem Cells. Cancer Res. 2010;70:9463–72. https://doi.org/10.1158/0008-5472.CAN-10-2388.
Greene SB, Herschkowitz JI, Rosen JM. The ups and downs of miR-205: Identifying the roles of miR-205 in mammary gland development and breast cancer. RNA Biol. 2010;7:300–4. https://doi.org/10.4161/rna.7.3.11837.
Avril-Sassen S, Goldstein LD, Stingl J, Blenkiron C, Le Quesne J, Spiteri I, et al. Characterisation of microRNA expression in post-natal mouse mammary gland development. BMC Genomics. 2009;10:548. https://doi.org/10.1186/1471-2164-10-548.
Feuermann Y, Robinson GW, Zhu BM, Kang K, Raviv N, Yamaji D, et al. The miR-17/92 cluster is targeted by STAT5 but dispensable for mammary development. Genesis. 2012;50:665–71. https://doi.org/10.1186/1471-2164-10-548.
Kahata K, Maturi V, Moustakas A. TGF-β Family Signaling in Ductal Differentiation and Branching Morphogenesis. Cold Spring Harb Perspect Biol. 2018;10:a031997. https://doi.org/10.1101/cshperspect.a031997.
Kohn EA, Du Z, Sato M, Van Schyndle CMH, Welsh MA, Yang Y an, et al. A novel approach for the generation of genetically modified mammary epithelial cell cultures yields new insights into TGFβ signaling in the mammary gland. Breast Cancer Res. 2010;12. https://doi.org/10.1186/bcr2728.
Michalak EM, Nacerddine K, Pietersen A, Beuger V, Pawlitzky I, Cornelissen-Steijger P, et al. Polycomb group gene Ezh2 regulates mammary gland morphogenesis and maintains the luminal progenitor pool. Stem Cells. 2013;31:1910–20. https://doi.org/10.1002/stem.1437.
Korkaya H, Paulson A, Charafe-Jauffret E, Ginestier C, Brown M, Dutcher J, et al. Regulation of mammary stem/progenitor cells by PTEN/Akt/beta-catenin signaling. PLoS Biol. 2009;7:e1000121. https://doi.org/10.1371/journal.pbio.1000121.
Lee J, Heo J, Kang H. miR-92b-3p-TSC1 axis is critical for mTOR signaling-mediated vascular smooth muscle cell proliferation induced by hypoxia. Cell Death Differ. 2019;26:1782–95. https://doi.org/10.1038/s41418-018-0243-z.
Liang G, Ling Y, Lin Q, Shi Y, Luo Q, Cen Y, et al. MiR-92b-3p Inhibits Proliferation of HER2-Positive Breast Cancer Cell by Targeting circCDYL. Front Cell Dev Biol. 2021;9. https://doi.org/10.3389/fcell.2021.707049.
Long M, Zhan M, Xu S, Yang R, Chen W, Zhang S, et al. miR-92b-3p acts as a tumor suppressor by targeting Gabra3 in pancreatic cancer. Mol Cancer. 2017;16. https://doi.org/10.1186/s12943-017-0723-7.
Ye Z, Shi J, Ning Z, Hou L, Hu CY, Wang C. MiR-92b-3p inhibits proliferation and migration of C2C12 cells. Cell Cycle. 2020;19:2906–17. https://doi.org/10.1080/15384101.2020.1827511.
Sengupta S, Nie J, Wagner RJ, Yang C, Stewart R, Thomson JA. MicroRNA 92b controls the G1/S checkpoint gene p57 in human embryonic stem cells. Stem Cells. 2009;27:1524–8. https://doi.org/10.1002/stem.84.
Miller DH, Sokol ES, Gupta PB. 3D Primary Culture Model to Study Human Mammary Development. Methods Mol Biol. 2017;1612:139–47. https://doi.org/10.1007/978-1-4939-7021-6_10.
Rosenbluth JM, Schackmann RCJ, Gray GK, Selfors LM, Li CMC, Boedicker M, et al. Organoid cultures from normal and cancer-prone human breast tissues preserve complex epithelial lineages. Nat Commun. 2020;11. https://doi.org/10.1038/s41467-020-15548-7.
Bonetti P, Climent M, Panebianco F, Tordonato C, Santoro A, Marzi MJ, et al. Dual role for miR-34a in the control of early progenitor proliferation and commitment in the mammary gland and in breast cancer. Oncogene. 2018;38:360–74. https://doi.org/10.1038/s41388-018-0445-3.
Bockmeyer CL, Christgen M, Müller M, Fischer S, Ahrens P, Länger F, et al. MicroRNA profiles of healthy basal and luminal mammary epithelial cells are distinct and reflected in different breast cancer subtypes. Breast Cancer Res Treat. 2011;130:735–45. https://doi.org/10.1007/s10549-010-1303-3.
Faridani OR, Abdullayev I, Hagemann-Jensen M, Schell JP, Lanner F, Sandberg R. Single-cell sequencing of the small-RNA transcriptome. Nat Biotechnol. 2016;34:1264–6. https://doi.org/10.1038/nbt.3701.
Acknowledgements
This work was supported in part by an unrestricted fund from the Harry M. Zweig Memorial Fund for Equine Research and by the Albert C. Bostwick Foundation to G. Van de Walle. These funding sources had no role in the study design, data collection, data analysis, data interpretation, writing of the report, or the decision to submit this manuscript for publication. We would like to thank following individuals for collecting and providing us with mammary tissue samples: Dorothy Supp, Erin Daugherity, Stephanie Pierson, Luce Guanzini, Patrick Burke, Dwight Hoffman, Michelle Delco, Joy Tomlinson, Amy Vasquez, Ed Rice, and Charles Danko.
Author information
Authors and Affiliations
Contributions
JM, MK, GR, PS and GVdW conceived and designed the study; JM, GR, and KB performed the wet-lab experiments; GR submitted samples for small RNA-seq; MK and PS analyzed the bioinformatics data; JM, MK, GR, KB, PS and GVdW interpreted the data; JM, GR and GVdW wrote the initial manuscript; MK and PS were major contributors in critically reviewing the manuscript. All authors reviewed this manuscript and approved the final version.
Corresponding author
Ethics declarations
Disclosure of Potential Conflicts of Interest
The authors declare that they have no conflicts of interest.
Research involving human participants and/or animals.
With exception of human breast tissue, all material used in this study was collected from animals after euthanasia or slaughter. Human tissue was obtained from discarded and de-identified breast tissue from reduction mammoplasty surgeries. No one involved in this study had any access to patient information, with exception of age and ethnicity.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
James L. Miller and Matt Kanke contributed equally to this work.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Miller, J.L., Kanke, M., Rauner, G. et al. Comparative Analysis of microRNAs that Stratify in vitro Mammary stem and Progenitor Activity Reveals Functionality of Human miR-92b-3p. J Mammary Gland Biol Neoplasia 27, 253–269 (2022). https://doi.org/10.1007/s10911-022-09525-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10911-022-09525-7