Abstract
A central goal of synthetic biology is the design of molecular controllers that can manipulate the dynamics of intracellular networks in a stable and accurate manner. To address the fact that detailed knowledge about intracellular networks is unavailable, integral-feedback controllers (IFCs) have been put forward for controlling molecular abundances. These controllers can maintain accuracy in spite of the uncertainties in the controlled networks. However, this desirable feature is achieved only if stability is also maintained. In this paper, we show that molecular IFCs can suffer from a hazardous instability called negative-equilibrium catastrophe (NEC), whereby all nonnegative equilibria vanish under the action of the controllers, and some of the molecular abundances blow up. We show that unimolecular IFCs do not exist due to a NEC. We then derive a family of bimolecular IFCs that are safeguarded against NECs when uncertain unimolecular networks, with any number of molecular species, are controlled. However, when IFCs are applied on uncertain bimolecular (and hence most intracellular) networks, we show that preventing NECs generally becomes an intractable problem as the number of interacting molecular species increases. NECs therefore place a fundamental limit to design and control of molecular networks.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
A main objective in synthetic biology is to control living cells [1,2,3,4,5,6]—a challenging problem that requires addressing a number of complicating factors displayed by intracellular networks:
- (N):
-
Nonlinearity. Intracellular networks are nonlinear (bimolecular), i.e. they include reactions involving two reacting molecules.
- (D):
-
Dimensionality. Intracellular networks are higher-dimensional, i.e. they contain a large number of coupled molecular species.
- (U):
-
Uncertainty. The experimental information about the structure, rate coefficients and initial conditions of intracellular networks is uncertain (incomplete).
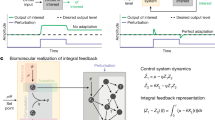
When embedded into an input network satisfying properties (N), (D) and (U), an ideal molecular controller network would ensure that the resulting output network autonomously traces a predefined dynamics in a stable and accurate manner over a desired time-interval. Controllers that maintain accuracy in spite of suitable uncertainties are said to achieve robust adaptation (homeostasis)—a fundamental design principle of living systems [7,8,9,10,11,12]. Control can be sought over deterministic dynamics when all of the molecular species are in higher-abundance [13, 15], or over stochastic dynamics when some species are present at lower copy-numbers [16,17,18]. See also Fig. 1, and Appendices A and B.
Schematic representation of biochemical control. A black-box input network \(\mathcal {R}_{\alpha }\) is displayed, consisting of unknown biochemical interactions shown in grey, where \(X_1 \rightarrow X_2\) (respectively, \(X_1 \dashv X_2\)) indicates that species \(X_1\) influences species \(X_2\) positively (respectively, negatively). The particular input network contains one interfacing species \(X_1\), shown as a green hexagon, that can be interfaced with a controller. The rest of the input species, that cannot be interfaced with a controller, are called residual species; one of the residual species, \(X_2\), is highlighted as a yellow triangle, while some of the other ones are displayed in grey. A controller network \(\mathcal {R}_{\beta , \gamma } = \mathcal {R}_{\beta } \cup \mathcal {R}_{\gamma }\) is also shown, consisting of controlling species \(Y_1\) and \(Y_2\) shown as a red circle and blue square, respectively, whose predefined biochemical interactions are shown in black. The controller consists of the core \(\mathcal {R}_{\beta } = \mathcal {R}_{\beta }(Y_1, Y_2)\), specifying how the controlling species interact among themselves, and the interface \(\mathcal {R}_{\gamma } = \mathcal {R}_{\gamma }(X_1, Y_1, Y_2)\), specifying how the controlling species \(Y_1\) and \(Y_2\) interact with the species \(X_1\). The composite network \(\mathcal {R}_{\alpha ,\beta ,\gamma } = \mathcal {R}_{\alpha } \cup \mathcal {R}_{\beta , \gamma }\) is called an output network. The particular controller \(\mathcal {R}_{\beta , \gamma }\) displayed corresponds to the network (8), with \(i = j = 1\), from Sect. 3 (Color figure online)
In context of electro-mechanical systems, accuracy robust to some uncertainties can be achieved via so-called integral-feedback controllers (IFCs) [19]. Loosely speaking, IFCs dynamically calculate a time-integral of a difference (error) between the target and actual values of the controlled variable. The error is then used to decrease (respectively, increase) the controlled variable when it deviates above (respectively, below) its target value via a negative-feedback loop. However, IFCs implementable with electro-mechanical systems are not necessarily implementable with biochemical reactions [20]. Central to this problem is the fact that the error takes both positive and negative values and, therefore, cannot be directly represented as a nonnegative molecular abundance. In this context, a non-biochemical IFC has been mapped to a bimolecular one in [21], which has been adapted in [22] and called the “antithetic integral-feedback controller" (AIFC).
Performance of the AIFC has been largely studied when unimolecular and/or lower-dimensional input networks are controlled [22,23,24,25]; in contrast, intracellular networks are generally bimolecular and higher-dimensional (challenges (N) and (D) stated above). For example, authors from [22] analyze performance of the AIFC in context of controlling average copy-numbers of intracellular species at the stochastic level. In this setting, in [22, Theorem 2], the authors specify a class of unimolecular input networks that can be controlled with the AIFC; in particular, to ensure stability, these input networks have to satisfy an algebraic constraint given as [22, Eq. 7]. This technical condition cannot generally be guaranteed to hold as it not only depends on the rate coefficients of the controller, but also on the uncertain rate coefficients of a given input network. More precisely, affine input networks, i.e. unimolecular input networks that contain one or more basal productions (zero-order reactions), can violate condition [22, Eq. 7]. In contrast, linear input networks, i.e. unimolecular networks with no basal production, always satisfy this condition. To showcase the performance of the AIFC, the authors from [22] put forward a gene-expression system as the input network, given by [22, Eq. 9], and demonstrate that the AIFC can arbitrarily control the average protein copy-number, and mitigate the uncertainties in the input rate coefficients (challenge (U) stated above). However, this unconditional success arises because basal transcription is not included in the linear gene-expression input network [22, Eq. 9], which ensures that condition [22, Eq. 7] always holds. A similar particular choice of an input network without basal production is put forward in [23], where the AIFC is experimentally implemented. The AIFC has also been analyzed in context of controlling species concentrations at the deterministic level in [24, 25]; however, the results are derived only for a restricted class of linear input networks that, due to lacking basal production, unconditionally satisfy [22, Theorem 2].
Questions of critical importance arise in context of controlling unimolecular networks: When the AIFC is applied on affine input networks (unimolecular networks with basal production), how likely is control to fail? Are the consequences of control failures biochemically safe or hazardous [26]? Does there exist a molecular IFC with a better stability performance than the AIFC? In particular, assume that a given intracellular network is modelled as being affine; then, the AIFC is successful provided the stability condition [22, Eq. 7] holds. However, a critical challenge arises because it is not possible to a-priori guarantee that [22, Eq. 7] holds, because the rate coefficients of the intracellular network are uncertain [challenge (U)].
While analyzing control of unimolecular networks in their own right is of some theoretical relevance, the vast majority of intracellular networks are not unimolecular [challenge (N)] and, hence, e.g. condition [22, Eq. 7] does not even apply. Therefore, a far more important question for intracellular control is: How do molecular IFCs perform when applied to bimolecular and higher-dimensional input networks? In particular, the structure of intracellular networks is itself in general uncertain; consequently, only approximate models can be put forward, that neglect a number of coupled molecular species and processes that are “hidden” in the intracellular system. Hence, even if a less-detailed model of an intracellular network happens to be successfully controlled, there is in general no guarantee that this remains true when a more-detailed model is used—a phenomenon we call phantom control.
The main objective of this paper is to address these questions. We show that at the center of these issues are equilibria—stationary solutions of the reaction-rate equations (RREs) that govern the deterministic dynamics of biochemical networks [13, 14]. In particular, molecular concentrations can reach only equilibria that are nonnegative. In this context, we show that IFCs can destroy all nonnegative equilibria of the controlled system and lead to a control failure; furthermore, this failure can be catastrophic, as some of the molecular concentrations can then experience an unbounded increase with time (blow-up). We call this hazardous phenomenon, involving absence of nonnegative equilibria and blow-up of some of the underlying species abundances, a negative-equilibrium catastrophe (NEC), which we outline in Fig. 2. To the best of our knowledge, analyses of NECs and the related challenges in molecular control, which are the focus of this paper, are absent from the literature. For example, the only form of instability presented in [22, 24, 25] are bounded deterministic oscillations, which average out at the stochastic level and do not correspond to violation of condition [22, Eq. 7]; in contrast, we show that this condition is violated when NECs occur.
Caricature representation of a successful and catastrophically failed intracellular control. a Displays a cell successfully controlled with the IFC in the setup shown in Fig. 1. The time-evolution of the underlying species concentrations are shown in b. In particular, the target species \(X_1\) approaches a desired equilibrium, shown as a black dashed line, and the equilibrium for the residual species \(X_2\) is positive. c Displays a cell that has taken lethal damage due to a failure of the IFC. In particular, as shown in d, the target equilibrium for \(X_1\) enforces a negative equilibrium for the residual species \(X_2\). However, since molecular concentrations are nonnegative, this equilibrium cannot be reached and, therefore, control fails. Furthermore, the failure is catastrophic, as concentrations of some of the underlying species (in this example, species \(X_2\) and \(Y_1\)) blow up, placing a lethal burden on the cell. b and d are obtained by solving the reaction-rate equations for the output network (16)\(\cup \)(19) from Sect. 5 with the dimensionless coefficients \((\alpha _0, \alpha _1, \alpha _2, \alpha _3) = (1, 1,1/10, 3/2)\), \((\beta _0, \beta _1, \gamma _1, \gamma _2, \gamma _3) = (100, 1, 10, 10, 1)\), and with \((\alpha _0, \alpha _1, \alpha _2, \alpha _3) = (1, 25,2/5,3/2)\), \((\beta _0, \beta _1, \gamma _1, \gamma _2, \gamma _3) = (100, 1, 10,2/3, 1)\), respectively
The paper is organized as follows. In Sects. 2–4, we present some fundamental results regarding control of unimolecular networks; these results are then used to facilitate the analysis of control of bimolecular networks, which we present in Sect. 5. In particular, in Sect. 2, we prove that unimolecular IFCs do not exist due to a NEC. We then derive a class of bimolecular IFCs given by (8) in Sect. 3; as a consequence of demanding in the derivation that the controlling variables are positive, we obtain IFCs that influence the target species both positively and negatively, in contrast to the AIFC that acts only positively. In Sect. 4, we apply different variants of these controllers on a unimolecular gene-expression network (9), and then generalize the results. In particular, we show that the AIFC can lead to a NEC when applied to (9), both deterministically and stochastically; more broadly, we show that the AIFC does not generically operate safely when applied to unimolecular networks. Furthermore, we prove that there exists a two-dimensional (two-species) IFC that eliminates NECs when applied to any (arbitrarily large) stable unimolecular input network. However, in Sect. 5 we demonstrate that, without detailed information about the input systems, NECs generally cannot be prevented when bimolecular networks are controlled. In particular, we show that, in stark contrast to dimension-independent control of unimolecular networks, control of bimolecular networks suffers from the curse of dimensionality - the problem becomes more challenging as the dimension of the input network increases. We conclude the paper by presenting a summary and discussion in Sect. 6. Notation and background theory are introduced as needed in the paper, and are summarized in Appendices A and B. Rigorous proofs of the results presented in Sects. 2, 4 and 5 are provided in Appendices C–D, E and F, respectively.
2 Nonexistence of unimolecular IFCs
In this section, we consider an arbitrary one-dimensional black-box input network \(\mathcal {R}_{\alpha } = \mathcal {R}_{\alpha }(X_1)\), where \(X_1\) is a single interfacing species and there are no residual species, and the goal is to control the dynamics of the target \(X_1\); see also Fig. 1 for a more general setup. In this paper, we assume all reaction networks are under mass-action kinetics [13, 14] with positive dimensionless rate coefficients, which are displayed above or below the reaction arrows; we denote the rate coefficients of \(\mathcal {R}_{\alpha }\) by \(\varvec{\alpha } \in \mathbb {R}_{>}^a\), where \(\mathbb {R}_{>}\) is the space of positive real numbers. In what follows, we say that a reaction network is unimolecular (respectively, bimolecular) if it contains at least one reaction with one (respectively, two) reactants, but no reaction with more reactants.
2.1 Non-biochemical controller
Let us consider an affine controller formally described by the network \(\mathcal {\bar{R}}_{\beta , \gamma } = \mathcal {\bar{R}}_{\beta }(\bar{Y}_1) \cup \mathcal {\bar{R}}_{\gamma }(X_1, \bar{Y}_1)\), given by
Here, \(\mathcal {\bar{R}}_{\beta } = \mathcal {\bar{R}}_{\beta }(\bar{Y}_1)\) is the controller core, describing the internal dynamics of the controlling species \(\bar{Y}_1\), where the reactant \(\varnothing \) denotes a source, while \(\mathcal {\bar{R}}_{\gamma } = \mathcal {\bar{R}}_{\gamma }(X_1, \bar{Y}_1)\) is the controller interface, specifying interactions between \(\bar{Y}_1\) and the target species \(X_1\) from the input network, see also Fig. 1. Let us denote abundances of species \(\{X_1, \bar{Y}_1\}\) from the output network \(\mathcal {R}_{\alpha } \cup \mathcal {\bar{R}}_{\beta , \gamma }\) at time \(t \ge 0\) by \((x_1, \bar{y}_1) = (x_1(t), \bar{y}_1(t)) \in \mathbb {R}^2\). At the deterministic level, formal reaction-rate equations (RREs) [13, 14] read
where \(f_1(x_1; \, \varvec{\alpha })\) is an unknown function describing the dynamics of \(\mathcal {R}_{\alpha }\), and \((x_1^*, \bar{y}_1^*) \in \mathbb {R}^2\) is the unique equilibrium of the output network, obtained by solving the RREs with zero left-hand sides. Assuming that \((x_1^*, \bar{y}_1^*)\) is globally stable, network (1) is an IFC; in particular, in this case, \(x_1^* = (\beta _0/\gamma _1)\) is independent of the initial conditions and the input coefficients \(\varvec{\alpha }\). However, controller (1) cannot be interpreted as a biochemical reaction network. In particular, the term \((-\gamma _{1} x_1)\) in (2) induces a process graphically described by \(X_1 \xrightarrow []{\gamma _1} X_1 - \bar{Y}_1\) in (1), which consumes species \(\bar{Y}_1\) even when its abundance is zero. Consequently, variables \((x_1, \bar{y}_1)\) may take negative values and, therefore, cannot be interpreted as molecular concentrations [20].
2.2 Unimolecular controllers
The only unimolecular analogue of the IFC (1), that contains only one controlling species \(Y_1\), is of the form
The RREs and the equilibrium for the output network \(\mathcal {R}_{\alpha } \cup \mathcal {R}_{\beta , \gamma }\) are given by
Given nonnegative initial conditions, variables \((x_1, y_1)\) from (4) are confined to the nonnegative quadrant \(\mathbb {R}_{\ge }^2\), and represent biochemical concentrations. However, the \(x_1\)-component of the equilibrium from (4) is negative and, therefore, not reachable by the controlled system. Furthermore, \(y_1\) is a monotonically increasing function of time, \(\textrm{d} y_1/\textrm{d} t > 0\), i.e. \(y_1\) blows up. We call this phenomenon a deterministic negative-equilibrium catastrophe (NEC), see also Appendix A. Network (3) not only fails to achieve control, but it introduces an unstable species and is, hence, biochemically hazardous. In Appendix C, we prove that a NEC occurs at both deterministic and stochastic levels for any candidate unimolecular IFC, which we state as the following theorem.
Theorem 2.1
There does not exist a unimolecular integral-feedback controller.
Proof
See Appendix C. \(\square \)
To the best of our knowledge, Theorem 2.1 has not been previously reported in the literature. A related result is presented in [23, Proposition S2.7] and states that a molecular controller \(\mathcal {R}_{\beta } \cup \mathcal {R}_{\gamma }\), satisfying a set of assumptions, including the assumption that the interface \(\mathcal {R}_{\gamma }\) contains only catalytic reactions, is a molecular IFC only if the core \(\mathcal {R}_{\beta }\) contains a bimolecular degradation. No such assumptions have been made in Theorem 2.1, which holds for all unimolecular networks; in particular, we allow interface \(\mathcal {R}_{\gamma }\) to contain non-catalytic reactions, such as \(Y_i \rightarrow X_j\) and \(X_i \rightarrow Y_j\).
3 Design of bimolecular IFCs
Theorem 2.1 implies that only bimolecular (and higher-molecular) biochemical networks may exert integral-feedback control. An approach to finding such networks is to map non-biochemical IFCs into biochemical networks, while preserving the underlying integral-feedback structure. This task can be achieved using special mappings called kinetic transformations [20]. Let us consider the non-biochemical system (2). The first step in bio-transforming (2) is to translate relevant trajectories \((x_1, \bar{y}_1)\) into the nonnegative quadrant. However, since \(\mathcal {R}_{\alpha }(X_1)\) is a black-box network, i.e. \(f_1(x_1; \, \varvec{\alpha })\) is unknown and unalterable, only \(\bar{y}_1\) can be translated; to this end, we define a new variable \(y_1 \equiv (\bar{y}_1 + T)\), with translation \(T > 0\), under which (2) becomes
Terms \((-\gamma _1 x_1)\) and \((-\gamma _2 T)\), called cross-negative terms [20], do not correspond to biochemical reactions and, therefore, must be eliminated. Let us note that cross-negative terms also play a central role in the questions of existence of other fundamental phenomena in biochemistry, such as oscillations, multistability and chaos [14, 20, 27]. Term \((-\gamma _1 x_1)\) can be eliminated with the so-called hyperbolic kinetic transformation, presented in Appendix D, which involves introducing an additional controlling species \(Y_2\) and extending system (5) into
Note that (5) and (6) have identical equilibria (time-independent solutions) for the species \(X_1\) and \(Y_1\), and that the equilibria for the species \(Y_1\) and \(Y_2\) have a hyperbolic relationship; furthermore, provided \(\beta _1\) is sufficiently large, time-dependent solutions of (5) and (6) are close as well, see Appendix D. On the other hand, cross-negative term \((-\gamma _2 T)\) can be eliminated via multiplication with \(x_1\) and any other desired factor; such operations do not influence the \(x_1\)-equilibrium, which is determined solely by the RREs for \(y_1\) and \(y_2\), and which we want to preserve. One option is to simply map \((-\gamma _2 T)\) to \((-\gamma _2 T x_1)\), and take T large enough to ensure that the \(y_1^*\)-equilibrium is positive; however, this approach requires the knowledge of \(f_1(x_1; \, \varvec{\alpha })\). A more robust approach is to map \((-\gamma _2 T)\) to \((-\gamma _2 T x_1 y_2)\), under which, defining \(\gamma _3 \equiv \gamma _2 T\), one obtains
The quadratic equation for \(y_1^*\) from (7) always has one positive solution; therefore, there always exists an equilibrium with positive \(y_1\)- and \(y_2\)-components.
In what follows, we largely consider input networks \(\mathcal {R}_{\alpha }(\mathcal {X})\) with at most two interfacing species \(\{X_1, X_2\}\), and focus on controlling the target species \(X_1\) with the bimolecular controllers induced by (7), given by
In particular, the controller core \(\mathcal {R}_{\beta }(Y_1, Y_2)\) consists of a production of \(Y_1\) from a source, and a bimolecular degradation of \(Y_1\) and \(Y_2\). On the other hand, the controller interface consists of the unimolecular reactions \(\mathcal {R}_{\gamma }^0(Y_2; \, X_1)\), and \(\mathcal {R}_{\gamma }^{+}(X_i; \, Y_1)\), that produce \(Y_2\) catalytically in \(X_1\), and \(X_i\) catalytically in \(Y_1\), respectively, and the bimolecular reaction \(\mathcal {R}_{\gamma }^{-}(X_j; \, Y_2)\) that degrades an interfacing species \(X_j\) catalytically in \(Y_2\). We call reactions \(\mathcal {R}_{\gamma }^{+}(X_i; \, Y_1)\) and \(\mathcal {R}_{\gamma }^{-}(X_j, Y_2)\) positive and negative interfacing, respectively. Furthermore, we say that positive (respectively, negative) interfacing is direct if \(i = 1\) (respectively, if \(j = 1\)), i.e. if it is applied to the target species \(X_1\); otherwise, the interfacing is said to be indirect. In Fig. 1, we display controller (8) with direct positive and negative interfacing applied to an input network with a single interfacing species \(X_1\).
As shown in this section, positive and negative interfacing arise together naturally when molecular IFCs are designed using the theoretical framework from [20]. In this context, it is interesting to note that the “housekeeping” sigma/anti-sigma system in E. coli, proposed to implement integral control [22], has been experimentally shown to be capable of exhibiting both positive and negative transcriptional control, at least when hijacked by bacteriophage [28]. On the other hand, while the AIFC from [22] is of the form (8), it lacks negative interfacing \(\mathcal {R}_{\gamma }^{-}(X_j; \, Y_2)\). In view of the derivation from this section, the AIFC is missing a key designing step, namely the translation from (5); consequently, NECs may occur due to \(y_1^*\)- and \(y_2^*\)-equilibria being negative. Let us note that the negative interfacing \(\mathcal {R}_{\gamma }^{-}(X_j; \, Y_2)\) has also been considered in [29], where this reaction is shown to be capable of eliminating oscillations at the deterministic level, and reducing variance at the stochastic level, for a particular gene-expression input network. In contrast, in this section, we have systematically derived reaction \(\mathcal {R}_{\gamma }^{-}(X_j; \, Y_2)\) in order to ensure that a positive equilibrium for \(Y_1\) and \(Y_2\) exists. Such matters are not discussed in [29], where basal transcription is set to zero in the gene-expression input network considered, so that negative equilibria are not encountered.
4 Control of unimolecular input networks
In this section, we study performance of the IFCs (8) when applied on unimolecular input networks. To this end, let us consider the input network \(\mathcal {R}_{\alpha }^1 = \mathcal {R}_{\alpha }^1(X_1, X_2)\), given by

We interpret (9) as a two-dimensional reduced (simplified) model of a higher-dimensional gene-expression network. In this context, \(X_1\) is a degradable protein species that is produced via translation from a degradable mRNA species \(X_2\), which is transcribed from a gene; some of the “hidden" species (dimensions), that are not explicitly modelled, such as genes, transcription factors and waste molecules, are denoted by \(\varnothing \). See also Fig. 3a for a schematic representation of network (9). The RREs of (9) have a unique globally stable equilibrium given by
For simplicity, in this section we assume that \(X_1\) is an interfacing species, while \(X_2\) is residual (i.e. \(X_2\) cannot be interfaced with a controller); the goal is to control the equilibrium concentration of the target species \(X_1\) at the deterministic level, and its average copy-number at the stochastic level. To this end, we embed different variants of the controller (8) into (9).
4.1 Pure positive interfacing
Let us first consider controller (8) with only positive interfacing, i.e. the AIFC from [22], which is denoted by \(\mathcal {R}_{\beta , \gamma }^{+} \equiv \mathcal {R}_{\beta } \cup \mathcal {R}_{\gamma }^0 \cup \mathcal {R}_{\gamma }^{+}\) and reads
The RREs for the output network (9)\(\cup \)(11) are given by
with the unique equilibrium
As anticipated in Sect. 3, the AIFC can lead to equilibria with negative \(y_1\)- and \(y_2\)-components. In particular, Eq. (13) implies that the output nonnegative equilibrium is destroyed when \((x_1^* = \beta _0/\gamma _1) < (x_1^{**} = \alpha _0 \alpha _2/(\alpha _1 \alpha _3))\). Hence, using only positive interfacing, it is not possible to achieve an output equilibrium below the input one. To determine the dynamical behavior of (9)\(\cup \)(11) when the nonnegative equilibrium ceases to exist, let us consider the linear combination of species concentration \((\alpha _3^{-1} x_1 + \alpha _1^{-1} \alpha _2 \alpha _3^{-1} x_2 + \gamma _1^{-1} (y_2 - y_1))\) that, using (12), satisfies
When \(\beta _0/\gamma _1 < \alpha _0 \alpha _2/(\alpha _1 \alpha _3)\), Eq. (14) implies that a species concentration blows up for all nonnegative initial conditions, i.e. the output network displays a deterministic NEC; by applying identical argument to the first-moment equations, it follows that a stochastic NEC occurs as well. This result is summarized as a bifurcation diagram in Fig. 4a.
Application of the IFCs (8) on the input network (9) with rate coefficients \((\alpha _1, \alpha _2, \alpha _3) = (2/5,2, 4)\), and piecewise constant \(\alpha _0 = \alpha _0(t)\) which changes at \(t = 50\) and leads to a catastrophic bifurcation. a Displays a bifurcation diagram for the output network (9)\(\cup \)(11), while b–d show the underlying deterministic and stochastic trajectories with \(\alpha _0 = 4\) for \(t < 50\) and \(\alpha _0 = 12\) for \(t \ge 50\), leading to a change in the parameter space indicated by the black arrow in a. The control coefficients are fixed to \((\beta _0, \beta _1,\gamma _1, \gamma _2) = (40, 4, 4, 4)\). Analogous plots are shown in e–h for the output network (9)\(\cup \)(15) with \(\alpha _0 = 12\) for \(t < 50\) and \(\alpha _0 = 4\) for \(t \ge 50\), and \((\beta _0, \beta _1,\gamma _1, \gamma _3) = (40, 4, 4, 4)\), and for the output network (9)\(\cup \)(16) in i–l using the same coefficient values as in a–d, and with \(\gamma _3 = 4\)
Let us consider network (9) with rate coefficients \(\varvec{\alpha } = (\alpha _0, \alpha _1, \alpha _2, \alpha _3)\) fixed so that the input \(x_1\)-equilibrium from (10) is given by \(x_1^{**} = 5\); then, the output \(x_1\)-equilibrium from (13) must satisfy the constraint \(x_1^* > 5\). Let us stress that, since the input rate coefficients \(\varvec{\alpha }\) (and the structure of the input network itself) are generally uncertain (see property (U) from Sect. 1), condition \(x_1^* > 5\) is not a-priori known. Assume the goal is to steer the output equilibrium to 10, i.e. we fix the control coefficients \(\beta _0\) and \(\gamma _1\) so that \(x_1^* = \beta _0/\gamma _1 = 10\); this setup is shown as a black dot at (5, 10) in Fig. 4a, which happens to lie in the region where the output network displays a nonnegative equilibrium. However, assume that at a future time, as a response to an environmental perturbation, an activating transcription factor binds to the underlying gene promoter, tripling the transcription rate of the input network, which we model by allowing the transcription rate coefficient \(\alpha _0\) to be time-dependent, see Fig. 3b. Such a perturbation would move the system from coordinate (5, 10) to (15, 10), into the unstable region, which we show as a black arrow in Fig. 4a. Put another way, the AIFC would fail at its main objective—maintaining accurate control robustly with respect to uncertainties.
In Fig. 4b–d, we show the deterministic and stochastic trajectories for the species \(X_1\), \(Y_1\) and \(Y_2\), obtained by solving the RREs (12) and applying the Gillespie algorithm [30] on (9)\(\cup \)(11), respectively; also shown as a dashed grey line in Fig. 4b is the target equilibrium \(x_1^* = 10\). For time \(t < 50\), when the input network operates as in Fig. 3a, and the output system is in the configuration (5, 10) from Fig. 4a, the AIFC achieves control. However, for time \(t \ge 50\), when the transcription rate has increased as in Fig. 3b, and the output system is at (15, 10) from Fig. 4a, control fails; even worse, the species \(Y_2\) blows up for all admissible initial conditions. Intuitively, this hazardous phenomenon (NEC) occurs because, when the target equilibrium is below the input one, the best accuracy result that the AIFC can achieve is to minimally increase \(X_1\). Such a task is accomplished with \(y_1 \rightarrow 0\) which, as a consequence of a hyperbolic relationship between \(y_1\) and \(y_2\), enforces \(y_2 \rightarrow \infty \), which is a worst stability result.
4.2 Pure negative interfacing
Consider controller (8) with pure direct negative interfacing, denoted by \(\mathcal {R}_{\beta , \gamma }^{-} \equiv \mathcal {R}_{\beta } \cup \mathcal {R}_{\gamma }^0 \cup \mathcal {R}_{\gamma }^{-}\) and given by
By analogous arguments as with controller (11), one can prove that deterministic and stochastic NECs occur when \(x_1^* > x_1^{**}\), i.e. controller (15) cannot steer the output equilibrium above the input one; when control above the input equilibrium is attempted, species \(Y_1\) blows up. We display a bifurcation diagram and trajectories for the output network (9)\(\cup \)(15) in Fig. 4e–h.
4.3 Combined positive and negative interfacing
Let us now analyze controller (8) with positive and negative interfacing applied together to the target species \(X_1\), as suggested by the derivation in Sect. 3. This controller is denoted by \(\mathcal {R}_{\beta , \gamma }^{\pm } \equiv \mathcal {R}_{\beta } \cup \mathcal {R}_{\gamma }^0 \cup \mathcal {R}_{\gamma }^{+} \cup \mathcal {R}_{\gamma }^{+}\) and given by
The RREs of the output network (9)\(\cup \)(16) have two equilibria, given by
where \(y_1^*\) satisfies
By design from Sect. 3, quadratic Eq. (18) always has exactly one positive equilibrium, so that no NEC can occur with controller (16); we confirm this fact in Fig. 4i–l.
4.4 Arbitrary unimolecular input networks
Test network (9) demonstrates that controller (8) with only positive, or only negative, interfacing does not generically ensure stability of the output network. In other words, the corresponding output network experiences NECs over a large region in the parameter space, as displayed in Fig. 4a and e. On the other hand, controller (8) with combined positive and negative interfacing, applied directly to the target species, induces no NEC, as shown in Fig. 4i. A special feature of unimolecular networks is that distinct species cannot influence each other negatively. Consequently, to ensure existence of a nonnegative equilibrium, negative interfacing must generally be applied directly to the target species, while positive interfacing can be applied directly or indirectly (i.e. to any suitable interfacing species). We now more formally state this result; for more details, see Appendix E. To aid the statement of the theorem, we say that an input network is degenerate if its \(x_1\)-equilibrium is zero, \(x_1^{**} = 0\); otherwise, the network is said to be nondegenerate. The set of all degenerate networks forms a negligibly small subset of general unimolecular networks.
Theorem 4.1
Consider an arbitrary nondegenerate unimolecular input network \(\mathcal {R}_{\alpha }\) whose RREs have an asymptotically stable equilibrium, the family of controllers \(\mathcal {R}_{\beta ,\gamma }\) given by (8), and the output network \(\mathcal {R}_{\alpha ,\beta ,\gamma } = \mathcal {R}_{\alpha } \cup \mathcal {R}_{\beta ,\gamma }\). Then, controller \(\mathcal {R}_{\beta ,\gamma }^{\pm }\) with both positive and negative interfacing, with negative interfacing being direct, ensures that the output network \(\mathcal {R}_{\alpha ,\beta ,\gamma }\) has a nonnegative equilibrium for all parameter values \((\varvec{\alpha }, \varvec{\beta },\varvec{\gamma }) \in \mathbb {R}_{>}^{a + b + c}\). On the other hand, the other variants of the controller (8) do not generically ensure that the output network \(\mathcal {R}_{\alpha ,\beta ,\gamma }\) has a nonnegative equilibrium; furthermore, when a nonnegative equilibrium does not exist, these controllers induce deterministic and stochastic blow-ups (NECs) for all nonnegative initial conditions.
Proof
See Appendix E. \(\square \)
Note that the only variant of (8) that generically eliminates NECs may be experimentally most challenging to implement: bimolecular reaction \(\mathcal {R}_{\gamma }^{-}\) must be applied directly to the target species.
In the degenerate case, when the equilibrium of the target species from the input network is zero, \(x_1^{**} = 0\), in addition to the positive–negative controller \(\mathcal {R}_{\beta ,\gamma }^{\pm }\), the AIFC \(\mathcal {R}_{\beta ,\gamma }^{+}\) also generically ensures existence of a nonnegative equilibrium. However, the degenerate input networks can describe only a small class of biochemical processes. For example, when there is no basal transcription, i.e. when \(\alpha _0 = 0\), the equilibrium of network (9) is zero and, consequently, the output equilibrium (13) is always nonnegative. In particular, the output equilibrium is then nonnegative independent of the uncertainties in the input coefficients, so that the key challenge (U) highlighted in Sect. 1 is mitigated. This gene-expression input network without basal transcription, \(\alpha _0 = 0\), has been used in [22] to demonstrate a desirable stochastic behavior of the AIFC. However, as we have shown in this section, when a more general gene-expression model is used, with \(\alpha _0 \ne 0\), the AIFC can fail and induce both deterministic and stochastic catastrophes as a consequence of the challenge (U). The results from [24, 25] also focus on similar degenerate input networks.
Let us stress that, while the AIFC is NEC-free when applied to unimolecular input networks with zero basal production, even an arbitrarily small basal production may cause a NEC to occur over a large parameter regime. For example, let us consider network (9) with the following rate coefficients: \(\alpha _0 = \varepsilon \), \(\alpha _2 = 1/\varepsilon ^2\) and \(\alpha _1 = \alpha _3 = 1\), where \(0 < \varepsilon \ll 1\) is sufficiently small, i.e. we are considering a slower-transcription and faster-translation gene-expression network [31]. It then follows from (13) that the AIFC can achieve control only when the target equilibrium is set to a sufficiently large value, \(\beta _0/\gamma _1 > \alpha _0 \alpha _2/(\alpha _1 \alpha _3) = 1/\varepsilon \); in other words, even though the basal transcription is small, nevertheless the AIFC fails catastrophically over a large parameter regime due to a large translation rate.
5 Control of bimolecular input networks: curse of dimensionality
As expressed by challenge (N) in Sect. 1, most intracellular networks are bimolecular, rather than unimolecular, limiting the applicability of Theorem 4.1. For example, in Sect. 4, we have used the unimolecular input network (9) with a time-dependent rate coefficient to model intracellular gene expression with regulated transcription. To obtain a more realistic model, instead of introducing an effective time-dependent rate coefficient, the reduced network (9) must be extended by including other coupled auxiliary species (e.g. transcription factors and genes) and processes (e.g. pre-transciption and post-translation events); the resulting extended input network is then bimolecular, and therefore Theorem 4.1 no longer applies. In particular, a special property of unimolecular networks is that distinct species can influence each other only positively; in contrast, distinct species can influence each other both positively and negatively in bimolecular networks. For this reason, when stable unimolecular input networks are controlled, NECs can be eliminated purely by ensuring that the controlling species equilibrium \(\textbf{y}^*\) is positive; the input species equilibrium \(\textbf{x}^*\) is then necessarily nonnegative. In other words, the problem of controlling unimolecular networks is independent of the challenge (D) from Sect. 1, i.e. the problem does not become more challenging as the dimension of the input network increases. On the other hand, we show in this section that, as a consequence of nonlinearities and positive–negative interactions among distinct species, the problem of controlling bimolecular networks suffers from the curse of dimensionality—the problem becomes more challenging as the dimension of the input network increases.
5.1 Two-species reduced input network: residual NEC
Let us consider a two-dimensional reduced model of an intracellular process, given by the bimolecular input network \(\mathcal {R}_{\alpha }^2(X_1, X_2)\) which reads
where \(X_1\) is produced from a source and converted into a degradable species \(X_2\) via first- and second-order conversion reactions. We assume that \(X_1\) is an interfacing and target species, while \(X_2\) is residual. The RREs of (19) have a unique asymptotically stable equilibrium, given by
where the function \(I_2 = I_2(x_1; \, \varvec{\alpha })\) is given by
We call (21) a residual invariant, which is simply the \(x_2\)-equilibrium expressed as a function of the \(x_1\)-equilibrium and the input coefficients \(\varvec{\alpha }\).
Application of the IFC (16) on the input network (19) with rate coefficients \((\alpha _0, \alpha _1, \alpha _3) = (200, 1/7, 5)\), and \(\alpha _2 = 1/3\) for \(t < 50\), which changes to \(\alpha _2 = 1\) for \(t \ge 50\) and leads to a catastrophic bifurcation. a–d Display the deterministic and stochastic trajectories for the output network (16)\(\cup \)(19), with control coefficients \((\beta _0, \beta _1,\gamma _1, \gamma _2, \gamma _3) = (100, 1,10,10,1)\)
Let us embed the controller (16) into (19), leading to the RREs of the output network given by
which display two equilibria, one of which is never nonnegative, while the other equilibrium satisfies
In particular, the functional form of the residual equilibrium \(x_2^{*}\) from (23) is the same as the form of \(x_2^{**}\) from (20); put another way, the form of the residual species equilibrium is invariant under control, justifying calling the function (21) a residual invariant. To ensure that the output network (16)\(\cup \)(19) displays a nonnegative equilibrium, the residual invariant (21), now evaluated at the target equilibrium \(x_1^* = \beta _0/\gamma _1\), must be nonnegative, giving rise to the condition
Therefore, while (16) guarantees existence of a nonnegative equilibrium for stable unimolecular input networks (see Theorem 4.1), the same is generally false for bimolecular networks, as the equilibrium of the residual species, which are not interfaced with the controller, can become negative.
In Fig. 5, we display the deterministic and stochastic trajectories for the output network (16)\(\cup \) (19) over a time-interval such that condition (24) is satisfied for \(t < 50\), and violated for \(t \ge 50\). One can notice that the output network undergoes deterministic and stochastic NECs. Critically, not only does the controlling species \(Y_1\) blow up, but also the residual species \(X_2\). In other words, controller (16) destabilizes the originally asymptotically stable input network (19). We call this hazardous phenomenon a residual NEC, as it arises because a residual species has no nonnegative equilibrium (equivalently, because a residual invariant is not nonnegative). Intuitively, when the concentration of the target species \(X_1\) is increased beyond the upper bound from (24), residual species \(X_2\), which influences \(X_1\) negatively, counteracts the positive action of the controlling species \(Y_1\), resulting in a joint blow-up. We also display this phenomenon in context of intracellular control in Fig. 2.
Let us stress that the condition \(I_2 \left( x_1^{*}, \varvec{\alpha } \right) \ge 0\) from (24) must be obeyed by every molecular controller (e.g. containing integral, proportional and/or derivative actions [19]) that cannot be interfaced with \(X_2\). Put another way, no matter how one chooses the functions \(g_1\), \(g_2\) and h in (22), the inequality \(I_2 \left( x_1^{*}, \varvec{\alpha } \right) \ge 0\) must be satisfied, which imposes an upper bound on the achievable output equilibrium via \(x_1^* < \alpha _3/\alpha _2\). The only way to eliminate this residual invariant condition is to eliminate the residual species \(X_2\), i.e. to design an appropriate controller that can be interfaced with both \(X_1\) and \(X_2\). However, as stated in challenges (D) and (U) in Sect. 1, intracellular networks generally contain a large number of coupled biochemical species with different biophysical properties, some of which may be unknown (hidden) or poorly characterized; therefore, it is generally unfeasible to demand that a controller is designed that can be interfaced with any desired species. In what follows, we further investigate this issue.
5.2 Three-species extended input network: phantom control
The two-dimensional network (19) has been put forward as a reduced model of an intracellular process, obtained by neglecting a number of molecular species that do not influence the dynamics of \(X_1\) and \(X_2\), or by using perturbation theory to eliminate slower or faster auxiliary species from considerations [32]. However, the goal of such model reductions is to capture the dynamics of the species \(X_1\) and \(X_2\) on a desired time-scale, and not necessarily to capture how the underlying higher-dimensional intracellular process responds to control. In this context, let us extend network (19) by including a “hidden” residual species \(X_3\) into consideration, which interacts with \(X_1\) and \(X_2\) according to the three-dimensional input network \(\mathcal {R}_{\alpha , \varepsilon }^3 = \mathcal {R}_{\alpha , \varepsilon }^3(X_1, X_2,X_3)\), given by
The residual species \(X_3\) influences \(X_1\) and \(X_2\) only weakly via the slower reaction \(X_3 \xrightarrow []{\varepsilon } X_1 + X_3\) in (25), where \(0 \le \varepsilon \ll 1\) is sufficiently small. One can readily show that the dynamics of the species \(X_1\) and \(X_2\) from the input networks (19) and (25) are identical as \(\varepsilon \rightarrow 0\), which we denote by writing \(\lim _{\varepsilon \rightarrow 0} \mathcal {R}_{\alpha , \varepsilon }^3 = \mathcal {R}_{\alpha }^2\). Furthermore, the RREs of the network (25) have a unique asymptotically stable positive equilibrium, given at the leading order by
where the residual invariants \(I_2 = I_2(x_1; \, \varvec{\alpha })\) and \(I_3 = I_3(x_1; \, \varvec{\alpha })\) are given by
In what follows, we let \(\varvec{\alpha } = (\alpha _0, \alpha _1, \alpha _2, \alpha _3, \alpha _4, \alpha _5, \alpha _6) = (200, 1/7, 1/3,5,1,4,1)\) and \(\varepsilon = 10^{-2}\); in Fig. 6, we demonstrate that the \((x_1, x_2)\)-dynamics of networks (19) and (25) are then close.
Input network (25) with rate coefficients \(\varvec{\alpha } = (\alpha _0, \alpha _1, \alpha _2, \alpha _3, \alpha _4, \alpha _5, \alpha _6) = (200, 1/7, 1/3,5,1,4,1)\) and different values of \(\varepsilon \). a–b display the deterministic trajectories for the species \(X_1\) and \(X_2\), respectively, from the input network (25) with \(\varepsilon = 0\) (equivalently, the input network (19)) and with \(\varepsilon = 10^{-2}\)
Let us now embed the controller (16) into (25); the RREs of the output network (16)\(\cup \)(25) have two equilibria, both of which have identical \((x_1, x_2, x_3)\)-components, given by
In addition to requiring that \(I_2(x_1^{*}; \, \varvec{\alpha }) \ge 0\), one must now also demand that \(I_3(x_1^{*}; \, \varvec{\alpha }) \ge 0\), to ensure that the (previously neglected) residual species \(X_3\) displays a nonnegative equilibrium, leading to
By accounting for the residual species \(X_3\), a lower bound is imposed on the achievable output equilibrium \(x_1^* = \beta _0/\gamma _1\) in (29), while no such lower bound is imposed in (24). Therefore, while the reduced network (19) is suitable to approximate the dynamics of \(X_1\) and \(X_2\) from the extended network (25), i.e. \(\lim _{\varepsilon \rightarrow 0} \mathcal {R}_{\alpha , \varepsilon }^3 = \mathcal {R}_{\alpha }^2\), network (19) is not suitable to approximate how (25) responds to control, i.e. \(\lim _{\varepsilon \rightarrow 0} (\mathcal {R}_{\alpha , \varepsilon }^3 \cup \mathcal {R}_{\beta , \gamma }^{\pm }) \ne (\mathcal {R}_{\alpha }^2 \cup \mathcal {R}_{\beta , \gamma }^{\pm })\). When a reduced network is successfully controlled under a parameter choice for which a corresponding extended network fails to be controlled, we say that a phantom control, as opposed to genuine control, occurs for the reduced network. Hence, when the lower bound in (29) is violated, network (16)\(\cup \)(19) displays a phantom control.
For the chosen input coefficients \(\varvec{\alpha }\), it follows from (29) that one can achieve the output equilibrium only within the small interval approximately given by \(13.6 \le x_1^* \le 15\); therefore, even small uncertainties in the input coefficients (challenge (U) from Sect. 1) can then move the system outside of this range, where the control fails. In Fig. 7a–c, we display the deterministic trajectories for the species \(X_1\), \(X_3\) and \(Y_2\) when the target equilibrium is given by \(x_1^* = \beta _0/\gamma _1 = 5\), thus violating only the lower bound from (29). One can notice that a deterministic NEC occurs—the target species \(X_1\) fails to reach the desired equilibrium, while the residual species \(X_3\) and the controlling species \(Y_2\) blow-up; one can similarly show that a stochastic NEC occurs. Analogous plots are shown in Fig. 7d–f when \(x_1^* = \beta _0/\gamma _1 = 30\), violating the upper bound from (29); one can notice that the species \(X_2\) and \(Y_1\) blow up, as in Fig. 5.
Application of the IFC (16) on the input network (25) with rate coefficients \((\alpha _0, \alpha _1, \alpha _2, \alpha _3, \alpha _4, \alpha _5, \alpha _6) = (200, 1/7, 1/3,5,1,4,1)\) and \(\varepsilon = 10^{-2}\). a–c Display some of the deterministic trajectories for the output network (16)\(\cup \)(25), when control coefficients are fixed to \((\beta _0, \beta _1,\gamma _1, \gamma _2, \gamma _3) = (50, 1,10,100,10)\). Analogous plots are shown in d–f when \((\beta _0, \beta _1,\gamma _1, \gamma _2, \gamma _3) = (300, 1,10,100,10)\)
5.3 Arbitrary bimolecular input networks
One can continue the model-refinement process which led from network (19) to (25) by including more auxiliary species \(X_4, X_5, X_6, \ldots \), each of which generally introduces an additional constraint, \(I_4, I_5, I_6, \ldots \ge 0\), which must be obeyed for an equilibrium to be nonnegative. More generally, let \(\mathcal {R}_{\alpha }\) be an arbitrary N-dimensional input network satisfying properties (N), (D) and (U) from Sect. 1, \(\mathcal {R}_{\beta , \gamma }\) an arbitrary M-dimensional IFC, and \(\mathcal {R}_{\alpha , \beta , \gamma } = \mathcal {R}_{\alpha } \cup \mathcal {R}_{\beta , \gamma }\) the corresponding \((N + M)\)-dimensional output network. In order to ensure that an equilibrium \((\textbf{x}^*, \textbf{y}^*) \in \mathbb {R}^{N + M}\) of \(\mathcal {R}_{\alpha , \beta , \gamma } \) is nonnegative, there are exactly two options.
5.3.1 Fine-tuning the control coefficients
The first option is to choose the control coefficients \(\varvec{\beta }\) and \(\varvec{\gamma }\) so that the equilibria of all of the \((N + M)\) species are nonnegative. However, this approach involves solving a system of \((N + M)\) nonlinear inequalities with uncertain coefficients \(\varvec{\alpha }\), which is an intractable theoretical problem. Furthermore, owing to a large number of inequalities, even when a nonnegative equilibrium does exist, the admissible values for \(\varvec{\beta }\) and \(\varvec{\gamma }\) may be confined to a small set, which can lead to a large parameter regime where IFCs can catastrophically fail. The fact that these issues are of concern when intracellular networks are controlled has been demonstrated already with as low as two-dimensional network (19) containing only one bimolecular reaction, and the three-dimensional network (25) containing two bimolecular reactions. Let us note that these issues are related to the fact that the proportion of the state-space \(\mathbb {R}^{N + M}\) occupied by the nonnegative orthant \(\mathbb {R}_{\ge }^{N + M}\) is given by \(2^{-(N + M)}\), which decreases exponentially as the dimension of the input network N increases—a fact known as the curse of dimensionality.
5.3.2 Interfacing the controller with every input species
A necessary condition to bypass the intractable problem of fine-tuning the control coefficients is to eliminate all of the residual invariant constraints \(I_n, I_{n+1}, \ldots \ge 0\), which can be achieved only by eliminating all of the residual species. Therefore, the second option is to design a suitable controller that can be interfaced with all of the N input species, which is an unfeasible experimental problem. However, for theoretical purposes, assume all the input species are interfacing; does there then exist an IFC that ensures a nonnegative equilibrium exists for every choice of the coefficients \(\varvec{\alpha }\), \(\varvec{\beta }\) and \(\varvec{\gamma }\), thus mitigating challenge (U)?
Theorem 5.1
Assume \(\mathcal {R}_{\alpha }\) is an arbitrary mass-action input network with N input species, all of which are interfacing. Then, there exists a bimolecular integral-feedback controller \(\mathcal {R}_{\beta , \gamma }\), containing 2N controlling species, such that the output network \(\mathcal {R}_{\alpha , \beta , \gamma } = \mathcal {R}_{\alpha } \cup \mathcal {R}_{\beta , \gamma }\) has a positive equilibrium for every choice of the rate coefficients \((\varvec{\alpha }, \varvec{\beta },\varvec{\gamma }) \in \mathbb {R}_{>}^{a + b + c}\).
Proof
See Appendix F for a constructive proof. \(\square \)
In Appendix F, we design a controller that achieves this task by generalizing the approach from Sect. 3, and applying the controller of the form (16) to every input species; therefore, the dimension of the controller scales with the dimension of the input network.
6 Discussion
In this paper, we have demonstrated that molecular IFCs can display severe stability issues when applied to biochemical networks subject to uncertainties. In particular, all nonnegative equilibria of the controlled network can vanish under IFCs, and some of the species abundances can then blow up. We call this hazardous phenomenon a negative-equilibrium catastrophe (NEC). In context of electro-mehanical systems, analogous phenomenon is known as integrator windup [19]—equilibria of the controlled system reach beyond the boundary of physically allowed values. For some electro-mechanical systems, one only requires the equilibria to be real (as opposed to complex); for biochemical systems, one additionally requires that the equilibria are also nonnegative. Let us stress that requiring an equilibrium to be nonnegative is significantly more restrictive than only requiring it is real. For example, while real linear systems of equations generically have a unique real solution, finding parameter regimes where a positive solution exists is non-trivial [33]; for nonlinear systems, determining such parameter regimes is generally even more challenging, see Sect. 5.3. The consequences of these issues, which are unavoidable for biochemical systems, have been under-explored in the molecular control literature to date.
We have shown in Sect. 2 that, due to the nonnegativity constraint, affine (unimolecular) biochemical systems cannot achieve integral control and, even worse, lead to catastrophes (NECs); in contrast, some affine electro-mechanical systems can achieve integral control [19]. In Sect. 3, using the theoretical framework from [20], we have then constructed a family of bimolecular (nonlinear) IFCs (8). In Sect. 4, we have proved in Theorem 4.1 that a particular two-dimensional (two-species) molecular IFC of the form (8) ensures existence of a nonnegative equilibrium when applied to stable input unimolecular networks of arbitrary dimensions; in particular, NECs can be eliminated in a dimension-independent manner when unimolecular networks are controlled. In contrast, in Sect. 5, we have demonstrated that control of bimolecular networks suffers from the curse of dimensionality—every species in the input network can in general introduce a constraint which must be obeyed for a nonnegative equilibrium to exist, leading to an intractable problem. For theoretical purposes, we have proved in Theorem 5.1 that, assuming all of the input species are known and interfacing—a generally experimentally unfeasible assumption, then there exists a higher-dimensional IFC that always eliminates NECs. Let us note that, in all of the biochemical networks studied in this paper, NECs simultaneously occur at both deterministic and stochastic levels. In particular, as opposed to the instability arising from bounded deterministic oscillations [22, 24, 25], which average out at the stochastic level, NECs in general persist in the stochastic setting.
Intracellular networks are in general bimolecular, higher-dimensional and subject to uncertainties, as respectively described by the properties (N), (D) and (U) in Sect. 1. Due to these challenges, generally only reduced (approximate) models of intracellular networks are available, which are obtained by eliminating a number of the underlying auxiliary coupled molecular species and reactions. The objective of these lower-dimensional reduced models is to capture the dynamics of desired intracellular species on a time-scale of interest [32]. However, using reduced models for purpose of control is in general unjustified due to NECs, i.e. reduced models do not necessarily capture how the underlying extended models respond to control. In particular, it takes only one of the many input species to display a negative equilibrium for control to fail and a catastrophic event to unfold; hence, including a previously neglected molecular species into a successfully controlled reduced model can result in an extended model for which control fails—a phenomenon we call phantom control, see Sect. 5. Let us note that, when a reduced model displaying a NEC is extended to include e.g. finite resources or dilution, then some of the underlying species concentrations, instead of growing to infinity, would reach finite, but large, values. Nevertheless, the effects of unwanted large concentrations, such as sequestration of ribosomes and depletion of metabolites, are potentially very harmful; moreover, in such a scenario, the underlying IFCs has failed to deliver control and perfect adaptation. NECs therefore place a fundamental limit to applicability of molecular IFCs in synthetic biology. In particular, if one attempts to address NECs, instead of a systematic approach, an ad-hoc approach is in general necessary, consisting of gathering detailed experimental information about a desired intracellular network and designing suitable higher-dimensional controllers that can be interfaced with appropriate input species.
References
D. Endy, Foundations for engineering biology. Nature 484, 449–453 (2005)
D. Del Vecchio, A.J. Dy, Y. Qian, Control theory meets synthetic biology. J. R. Soc. Interface 13(120), 3–43 (2016)
T.S. Gardner, C.R. Cantor, J.J. Collins, Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342 (2000)
M.B. Elowitz, S. Leibler, A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338 (2000)
J. Chappell, M.K. Takahashi, J.B. Lucks, Creating small transcription activating RNAs. Nat. Chem. Biol. 11(3), 214–220 (2015)
F.J. Isaacs, D.J. Dwyer, C. Ding, D.D. Pervouchine, C.R. Cantor, J.J. Collins, Engineered riboregulators enable post-transcriptional control of gene expression. Nat. Biotechnol. 22(7), 841–847 (2004)
T. Drengstig, H.R. Ueda, P. Ruoff, Predicting perfect adaptation motifs in reaction kinetic networks. J. Phys. Chem. B 112(51), 16752–16758 (2008)
J.E. Ferrell, Perfect and near-perfect adaptation in cell signaling. Cell Syst. 2(2), 62–67 (2016)
F.A. Chandra, G. Buzi, J.C. Doyle, Glycolytic oscillations and limits on robust efficiency. Science 333(6039), 187–192 (2011)
N. Barkai, S. Leibler, Robustness in simple biochemical networks. Nature 387, 913–917 (1997)
P. Spiro, J. Parkinson, H.G. Othmer, A model of excitation and adaptation in bacterial chemotaxis. PNAS 94, 7263–7268 (1997)
T.M. Yi, Y. Huang, M.I. Simon, J. Doyle, Robust perfect adaptation in bacterial chemotaxis through integral feedback control. PNAS 97(9), 4649–4653 (2000)
M. Feinberg, Foundations of Chemical Reaction Network Theory (Springer, Berlin, 2019)
P. Érdi, J. Tóth, Mathematical Models of Chemical Reactions. Theory and Applications of Deterministic and Stochastic Models (Manchester University Press, Princeton University Press, Princeton, 1989)
T.G. Kurtz, The relationship between stochastic and deterministic models for chemical reactions. J. Chem. Phys. 57, 2976–2978 (1972)
R. Erban, J. Chapman, Stochastic Modelling of Reaction-Diffusion Processes. Cambridge Texts in Applied Mathematics (Cambridge University Press, Cambridge, 2019)
S. Kar, W.T. Baumann, M.R. Paul, J.J. Tyson, Exploring the roles of noise in the eukaryotic cell cycle. Proc. Natl. Acad. Sci. USA 106, 6471–6476 (2009)
J.M.G. Vilar, H.Y. Kueh, N. Barkai, S. Leibler, Mechanisms of noise-resistance in genetic oscillators. PNAS 99(9), 5988–5992 (2002)
K.J. Åström, T. Hägglund, PID Controllers: Theory, Design, and Tuning (Instrument Society of America, Pittsburgh, 1995)
T. Plesa, T. Vejchodský, R. Erban, Chemical reaction systems with a homoclinic bifurcation: an inverse problem. J. Math. Chem. 54(10), 1884–1915 (2016)
K. Oishi, E. Klavins, Biomolecular implementation of linear I/O systems. IET Syst. Biol. 5(4), 252–260 (2011)
C. Briat, A. Gupta, M. Khammash, Antithetic integral feedback ensures robust perfect adaptation in noisy bimolecular networks. Cell Syst. 2(1), 15–26 (2016)
S.K. Aoki, G. Lillacci, A. Gupta, A. Baumschlager, D. Schweingruber, M. Khammash, A universal biomolecular integral feedback controller for robust perfect adaptation. Nature 570, 533–537 (2019)
N. Olsman, A.A. Baetica, F. Xiao, Y.P. Leong, J. Doyle, R. Murray, Hard limits and performance tradeoffs in a class of antithetic integral feedback networks. Cell Syst. 9(1), 49–63 (2019)
N. Olsman, F. Xiao, J. Doyle, Architectural principles for characterizing the performance of antithetic integral feedback networks. ISince 14, 277–291 (2019)
A. Boo, T. Ellis, G.B. Stan, Host-aware synthetic biology. Current Opin. Syst. Biol. 14, 66–72 (2019)
T. Plesa, T. Vejchodský, R. Erban, Test Models for Statistical Inference: Two-Dimensional Reaction Systems Displaying Limit Cycle Bifurcations and Bistability (Stochastic Dynamical Systems, Multiscale Modeling, Asymptotics and Numerical Methods for Computational Cellular Biology) (2017)
U.K. Sharma, D. Chatterji, Differential mechanisms of binding of anti-sigma factors Escherichia coli Rsd and bacteriophage T4 AsiA to E. coli RNA polymerase lead to diverse physiological consequences. J. Bacteriol. 190, 3434–3443 (2008)
A. Gupta, M. Khammash, An antithetic integral rein controller for bio-molecular networks. IEEE 58th Conference on Decision and Control (CDC), Nice, France: 2808–2813 (2019)
D.T. Gillespie, Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 81(25), 2340–2361 (1977)
J. Hausser, A. Mayo, L. Keren, U. Alon, Central dogma rates and the trade-off between precision and economy in gene expression. Nat. Commun. (2019). https://doi.org/10.1038/s41467-018-07391-8
G.A. Pavliotis, A.M. Stuart, Multiscale Methods: Averaging and Homogenization (Springer, New York, 2008)
L. Dines, On positive solutions of a system of linear equations. Ann. Math. 28(1/4), 386–392 (1926)
D. Cox, J. Little, D. O’Shea, Using Algebraic Geometry, second. (Springer, Cham, 2005)
M. Fiedler, V. Pták, On matrices with non-positive off-diagonal elements and positive principal minors. Czechoslov. Math. J. 12(3), 382–400 (1962)
L. Farina, S. Rinaldi, Positive Linear Systems: Theory and applications (Wiley, Hoboken, 2000). https://doi.org/10.1002/9781118033029
D.T. Gillespie, A rigorous derivation of the chemical master equation. Physica A 188(1), 404–425 (1992)
N.G. Van Kampen, Stochastic Processes in Physics and Chemistry (Elsevier, Amsterdam, 2007)
T. Plesa, G.B. Stan, T.E. Ouldridge, W. Bae, Quasi-robust control of biochemical reaction networks via stochastic morphing. J. R. Soc. Interface (2021). https://doi.org/10.1098/rsif.2020.0985
W. Klonowski, Simplifying principles for chemical and enzyme reaction kinetics. Biophys. Chem. 18(3), 73–87 (1983)
Funding
This work was supported by the EPSRC Grant EP/P02596X/1. Thomas E. Ouldridge would like to thank the Royal Society for a University Research Fellowship.
Author information
Authors and Affiliations
Contributions
TP, AD and TEO concetualized the study; TP performed the mathematical analyses and simulations, and wrote the paper; TEO secured the funding.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendices
Appendix A: Background
1.1 Notation
Given sets \(\mathcal {A}_1\) and \(\mathcal {A}_2\), their union, intersection, and difference are denoted by \(\mathcal {A}_1 \cup \mathcal {A}_2\), \(\mathcal {A}_1 \cap \mathcal {A}_2\), and \(\mathcal {A}_1 {\setminus } \mathcal {A}_2\), respectively. The largest element of a set of numbers \(\mathcal {A}\) is denoted by \(\max \mathcal {A}\). The empty set is denoted by \(\emptyset \). Set \(\mathbb {Z}\) is the space of integer numbers, \(\mathbb {Z}_{\ge }\) the space of nonnegative integer numbers, and \(\mathbb {Z}_{>}\) the space of positive integer numbers. Similarly, \(\mathbb {R}\) is the space of real numbers, \(\mathbb {R}_{\ge }\) the space of nonnegative real numbers, and \(\mathbb {R}_{>}\) the space of positive real numbers. Euclidean column vectors are denoted in boldface, \(\textbf{x} = (x_1, x_2, \ldots , x_N)^{\top } \in \mathbb {R}^{N} = \mathbb {R}^{N \times 1}\), where \(\cdot ^{\top }\) denotes the transpose operator. The i-standard basis vector is denoted by \(\textbf{e}_i \equiv (\delta _{i,1}, \delta _{i,2}, \ldots , \delta _{i,N})^{\top } \in \mathbb {R}^N\), where \(\delta _{i,j} = 0\) if \(i \ne j\), and \(\delta _{i,j} = 1\) if \(i = j\). The zero-vector is denoted by \(\textbf{0} \equiv (0, 0, \ldots , 0)^{\top } \in \mathbb {R}^N\), and we let \((\textbf{1} \equiv \sum _{i = 1}^N \textbf{e}_i) \in \mathbb {R}^N\). Given two Euclidean vectors \(\textbf{x}, \textbf{y} \in \mathbb {R}^{N}\), their inner-product is denoted by \(\langle \textbf{x}, \textbf{y} \rangle \equiv \sum _{i = 1}^N x_i y_i\). Abusing the notation slightly, given two sequences \(u, v: \mathbb {Z}_{\ge }^N \rightarrow \mathbb {R}\), their inner-product is also denoted by \(\langle u, v \rangle \equiv \sum _{\textbf{x} \in \mathbb {Z}_{\ge }^N} u(\textbf{x}) v(\textbf{x})\); we let \(\Vert u \Vert _{l_1} \equiv \sum _{\textbf{x} \in \mathbb {Z}_{\ge }^N} |u(\textbf{x})|\) denote the \(l^1\)-norm of u. Given a matrix \(A \in \mathbb {R}^{N \times N}\), with (i, j)-element \(\alpha _{i,j} \in \mathbb {R}\), we denote the i-row and j-column of A by \(\varvec{\alpha }_{i, \cdot } \equiv (\alpha _{i,1}, \alpha _{i,2}, \ldots , \alpha _{i,N}) \in \mathbb {R}^{1 \times N}\), and \(\varvec{\alpha }_{\cdot , j} \equiv (\alpha _{1,j}, \alpha _{2,j}, \ldots , \alpha _{N,j})^{\top } \in \mathbb {R}^{N \times 1}\), respectively. The identity matrix is denoted by \(I \equiv (\textbf{e}_1, \textbf{e}_2, \ldots , \textbf{e}_N) \in \mathbb {R}^{N \times N}\), the zero matrix by \(0 \equiv (\textbf{0}, \textbf{0}, \ldots , \textbf{0}) \in \mathbb {R}^{N \times N}\), and diagonal matrices are denoted by \(\text {diag}(\alpha _{1,1}, \alpha _{2,2}, \ldots , \alpha _{N,N}) \equiv (\alpha _{1,1} \textbf{e}_1, \alpha _{2,2} \textbf{e}_2, \ldots , \alpha _{N, N} \textbf{e}_N)\in \mathbb {R}^{N \times N}\). Determinant of a matrix A is denoted by |A|.
1.2 Appendix A.1: Biochemical reaction networks
Let \(\mathcal {R}_{\alpha } = \mathcal {R}_{\alpha }(\mathcal {X})\) be a reaction network describing interactions, under mass-action kinetics, between N biochemical species \(\mathcal {X} = \{X_1, X_2, \ldots , X_N\}\) in a well-mixed unit-volume reactor [13], as specified by the following a reactions:
Here, \(\varvec{\alpha } = (\alpha _1, \alpha _2, \ldots , \alpha _a) \in \mathbb {R}_{>}^a\) are the positive rate coefficients of the reactions from \(\mathcal {R}_{\alpha }\). Nonnegative vectors \(\varvec{\nu }_{j, \cdot } = (\nu _{j, 1}, \nu _{j,2}, \ldots , \nu _{j,N})^{\top } \in \mathbb {Z}_{\ge }^N\) and \(\varvec{\bar{\nu }}_{j, \cdot } = (\bar{\nu }_{j, 1}, \bar{\nu }_{j,2}, \ldots , \bar{\nu }_{j,N})^{\top } \in \mathbb {Z}_{\ge }^N\) are the reactant and product stoichiometric coefficients of the j-reaction, respectively. If \(\varvec{\nu }_{j, \cdot } = \textbf{0}\) (respectivelty, \(\varvec{\bar{\nu }}_{j, \cdot } = \textbf{0}\)), then the reactant (respectively, product) of the j-reaction is the null-species, denoted by \(\varnothing \); such reactions involve biochemical species that are not explicitly modelled, or represent non-biochemical processes. When convenient, we denote two irreversible reactions \((\sum _{l = 1}^N \nu _{i, l} X_l \xrightarrow []{\alpha _{i}} \sum _{l = 1}^N \bar{\nu }_{i, l} X_l) \in \mathcal {R}_{\alpha }\) and \((\sum _{l = 1}^N \bar{\nu }_{i, l} X_l \xrightarrow []{\alpha _{j}} \sum _{l = 1}^N \nu _{i, l} X_l) \in \mathcal {R}_{\alpha }\) jointly as the single reversible reaction  . Species \(X_i\) is a catalyst in the j-reaction from \(\mathcal {R}_{\alpha }\) if \(\nu _{j, i} = \bar{\nu }_{j, i} \ne 0\); if \(X_i\) is a catalyst in all of the reaction from \(\mathcal {R}_{\alpha }\), then we write \(\mathcal {R}_{\alpha } = \mathcal {R}_{\alpha }(\mathcal {X} {\setminus } X_i; \, X_i)\). The order of the j-reaction from network \(\mathcal {R}_{\alpha }\) is given by \(\langle \textbf{1}, \varvec{\nu }_{j, \cdot } \rangle \in \mathbb {Z}_{\ge }\). The order of reaction network \(\mathcal {R}_{\alpha }\) is given by \(\text {max} \{\langle \textbf{1}, \varvec{\nu }_{j, \cdot } \rangle | j \in \mathcal {A}\}\); first-order (respectively, second-order) reaction networks are also said to be unimolecular (respectively, bimolecular).
. Species \(X_i\) is a catalyst in the j-reaction from \(\mathcal {R}_{\alpha }\) if \(\nu _{j, i} = \bar{\nu }_{j, i} \ne 0\); if \(X_i\) is a catalyst in all of the reaction from \(\mathcal {R}_{\alpha }\), then we write \(\mathcal {R}_{\alpha } = \mathcal {R}_{\alpha }(\mathcal {X} {\setminus } X_i; \, X_i)\). The order of the j-reaction from network \(\mathcal {R}_{\alpha }\) is given by \(\langle \textbf{1}, \varvec{\nu }_{j, \cdot } \rangle \in \mathbb {Z}_{\ge }\). The order of reaction network \(\mathcal {R}_{\alpha }\) is given by \(\text {max} \{\langle \textbf{1}, \varvec{\nu }_{j, \cdot } \rangle | j \in \mathcal {A}\}\); first-order (respectively, second-order) reaction networks are also said to be unimolecular (respectively, bimolecular).
Given a class of biochemical reaction networks, parametrized by the underlying rate coefficients, it may be of interest if a given property is likely to be true when all admissible values of the rate coefficients are considered, which motivates the following definition. In what follows, sets whose total size is arbitrarily small are said to be of measure zero.
Definition A.1
(Genericity) Consider a mass-action reaction network \(\mathcal {R}_{\alpha }\) parametrized by the rate coefficients \(\varvec{\alpha } \in \mathbb {S}_{\alpha }\), where \(\mathbb {S}_{\alpha } \subset \mathbb {R}_{>}^a\) is a nonempty open set. Assume \(\mathbb {S}_{\alpha }\) is partitioned according to \(\mathbb {S}_{\alpha } = \Omega _{\alpha } \cup \omega _{\alpha }\), with \(\Omega _{\alpha } \cap \omega _{\alpha } = \emptyset \), where \(\omega _{\alpha }\) is a set of measure zero. A property is said to be generic for network \(\mathcal {R}_{\alpha }\) and set \(\mathbb {S}_{\alpha }\) if it holds for all \(\varvec{\alpha } \in \Omega _{\alpha }\) and fails to hold for all \(\varvec{\alpha } \in \omega _{\alpha }\).
Example Empy set \(\omega _{\alpha } = \emptyset \), and set \(\omega _{\alpha } = \{\varvec{\alpha }_1, \varvec{\alpha }_2, \ldots \}\), containing finitely or countably infinitely many points, have measure zero. All the points \(\varvec{\alpha }\) where a non-trivial polynomial \(\mathcal {P}(\varvec{\alpha })\) vanishes is also a set of measure zero [34]; for example, given a nonzero matrix \(A = A(\varvec{\alpha })\), with (i, j)-element \(\alpha _{i,j}\), the set of all points \(\varvec{\alpha }\) such that \(|A(\varvec{\alpha })| = 0\) has zero measure.
1.3 Appendix A.2: Dynamical models of reaction networks
In what follows, we present deterministic and stochastic models of mass-action reaction networks, and provide definitions in context of blow-ups.
1.3.1 Appendix A.2.1: Deterministic model
Let \(\textbf{x}(t; \, \varvec{\alpha }) = (x_1(t; \, \varvec{\alpha }), x_2(t; \, \varvec{\alpha }), \ldots , x_N(t; \, \varvec{\alpha }))^{\top } \in \mathbb {R}_{\ge }^N\) be a concentration vector at time \(t \in \mathbb {R}_{\ge }\) for the species \(\mathcal {X} = \{X_1, X_2, \ldots , X_N\}\) from the network \(\mathcal {R}_{\alpha }\), given by (30). A deterministic model of the network \(\mathcal {R}_{\alpha }\) describes the time-evolution of \(\textbf{x} = \textbf{x}(t; \, \varvec{\alpha })\) with a system of first-order ordinary differential equations (ODEs), called the reaction-rate equations (RREs) [13, 14, 16], given by
where \(\Delta \textbf{x}_{j, \cdot } \equiv (\bar{\varvec{\nu }}_{j, \cdot } - \varvec{\nu }_{j, \cdot }) \in \mathbb {Z}^N\) is the reaction vector of the j-reaction, and \(\textbf{x}^{\varvec{\nu _{j,\cdot }}} \equiv \prod _{i = 1}^N x_i^{\nu _{j,i}}\) with \(0^0 \equiv 1\). Function \(\varvec{\mathcal {K}}(\cdot ; \, \varvec{\alpha }): \mathbb {R}^N \rightarrow \mathbb {R}^N\), called a kinetic function (see also Appendix D), is a polynomial of degree \(m = \text {max} \{\langle \textbf{1}, \varvec{\nu }_{j, \cdot } \rangle | j \in \mathcal {A}\}\) in \(\textbf{x}\), which we denote by \(\varvec{\mathcal {K}}(\textbf{x}; \, \varvec{\alpha }) \in \mathbb {P}_{m}(\mathbb {R}^N; \, \mathbb {R}^N)\). Vector \(\textbf{x}^* = \textbf{x}^*(\varvec{\alpha }) < \varvec{\infty }\) is called an equilibrium of the RREs (31) if \(\varvec{\mathcal {K}}(\textbf{x}^*; \, \varvec{\alpha }) = \textbf{0}\).
We now present an important property of the RREs.
Theorem A.1
The nonnegative orthant \(\mathbb {R}_{\ge }^N\) is an invariant set for the ODEs (31).
Proof
See [13]. \(\square \)
In this paper, unimolecular networks are of interest and, to this end, we introduce the following definition.
Definition A.2
(Cross-nonnegative matrix) A matrix \(A \in \mathbb {R}^{N \times N}\) with nonnegative off-diagonal elements, \(\alpha _{i,j} \ge 0\) for all \(i,j \in \{1,2, \ldots , N\}\) such that \(i \ne j\), is said to be cross-nonnegative.
Remark
Cross-nonnegative matrices are known as negative Z, quasi-positive, essentially nonnegative, and Metzler matrices in the literature [35, 36].
In context of unimolecular reaction networks, Theorem A.1 implies the following corollary.
Corollary A.1
For unimolecular reaction networks, the kinetic function from the RREs (31) is given by \(\varvec{\mathcal {K}}(\textbf{x}; \, \varvec{\alpha }) = (\varvec{\alpha }_{\cdot ,0} + A \textbf{x})\), where \(\varvec{\alpha }_{\cdot ,0} = (\alpha _{1,0}, \alpha _{2,0}, \ldots , \alpha _{N,0})^{\top } \in \mathbb {R}_{\ge }^N\) is a nonnegative vector, while \(A \in \mathbb {R}^{N \times N}\) is a cross-nonnegative matrix.
A linear ODE system \(\textrm{d}\textbf{x}/\textrm{d} t = (\varvec{\alpha }_{\cdot ,0} + A \textbf{x})\) is said to be asymptotically stable (also simply referred to as stable in this paper) if all the eigenvalues of A have negative real parts.
1.3.2 Appendix A.2.2: Stochastic model
Let \(\textbf{X}(t; \varvec{\alpha }) = (X_1(t; \varvec{\alpha }), X_2(t; \varvec{\alpha }), \ldots , X_N(t; \varvec{\alpha }))^{\top } \in \mathbb {Z}_{\ge }^N\) be a copy-number vector at time \(t \in \mathbb {R}_{\ge }\) for the species \(\mathcal {X} = \{X_1, X_2, \ldots , X_N\}\) from the network \(\mathcal {R}_{\alpha }\), given by (30); abusing the notation slightly, we denote the points in the state-space for \(\textbf{X}(t; \varvec{\alpha })\) using the same symbol as the concentration vector from (31), i.e. by \(\textbf{x} = (x_1, x_2, \ldots , x_N)^{\top } \in \mathbb {Z}_{\ge }^N\). A stochastic model of the reaction network \(\mathcal {R}_{\alpha }\) describes the time-evolution of \(\textbf{X} = \textbf{X}(t; \varvec{\alpha })\) as a continuous-time discrete-space Markov chain [16, 37] characterized via a difference-operator \(\mathcal {L}_{\alpha }\), called the generator, given by [38]
where \(u: \mathbb {Z}_{\ge }^N \rightarrow \mathbb {R}\) belongs to a suitable function space. Here, \(\Delta \textbf{x}_{j, \cdot } = (\bar{\varvec{\nu }}_{j, \cdot } - \varvec{\nu }_{j, \cdot }) \in \mathbb {Z}^N\) is the j-reaction vector, while \(\textbf{x}^{\underline{\varvec{\nu }_{j,\cdot }}} = \prod _{i = 1}^N x_i^{\underline{\nu _{j,i}}}\), with \(x_i^{\underline{\nu _{j,i}}} = x_i (x_i - 1) \ldots (x_i - \nu _{j,i} + 1)\) and \(x^{\underline{0}} \equiv 1\) for all \(x \in \mathbb {Z}_{\ge }\). Furthermore, \(E_{\textbf{x}}^{\Delta \textbf{x}_{j,\cdot }} = \prod _{i = 1}^N E_{x_i}^{\Delta x_{j,i}}\) is a step-operator such that \(E_{\textbf{x}}^{\Delta \textbf{x}_{j,\cdot }} u(\textbf{x}) = u(\textbf{x} + \Delta \textbf{x}_{j,\cdot })\).
Let \(p(\cdot ,t; \, \varvec{\alpha }): \mathbb {Z}_{\ge }^N \rightarrow [0,1]\) be the probability-mass function (PMF) at time t of the Markov chain with generator (32), and let \(f_{\textbf{x}}: \mathbb {Z}_{\ge }^N \rightarrow \mathbb {R}\) be a suitable function of the species copy-numbers. We let \(\mathbb {E} f_{\textbf{x}} = \mathbb {E} f_{\textbf{x}}(t; \, \varvec{\alpha }) \equiv \langle f_{\textbf{x}}(\cdot ), p(\cdot , t; \, \varvec{\alpha }) \rangle \) denote the expectation of \(f_{\textbf{x}}(\textbf{x})\) at time t with respect to \(p(\textbf{x},t; \, \varvec{\alpha })\). In this paper, we focus on the average species copy-numbers, i.e. on the first-moment vector \(\mathbb {E} \textbf{X} = \mathbb {E} \textbf{X}(t; \, \varvec{\alpha }) = (\mathbb {E} X_1(t; \, \varvec{\alpha }), \mathbb {E} X_2(t; \, \varvec{\alpha }), \ldots , \mathbb {E} X_N(t; \, \varvec{\alpha }))^{\top } \in \mathbb {R}_{\ge }^N\), which evolves in time according to the ODEs [16, 38]
In the special case of unimolecular reaction networks, the first-moment Eqs. (33) and the RREs (31) are formally equivalent; more generally, the less-detailed deterministic and the more-detailed stochastic models match in the thermodynamic limit [15].
1.3.3 Appendix A.2.3: Blow-up and negative-equilibrium catastrophe
In this paper, we focus on the circumstances when some of the species abundance experiences an unbounded growth, and introduce the following definition for this purpose.
Definition A.3
(Blow-up) Reaction network \(\mathcal {R}_{\alpha }(\mathcal {X})\) is said to blow up deterministically for a given initial condition if \(\lim _{t \rightarrow \infty } x_i(t; \, \varvec{\alpha }) = \infty \) for some \(i \in \{1, 2, \ldots , N\}\), where the species concentration \(\textbf{x}(t; \, \varvec{\alpha }) \in \mathbb {R}_{\ge }^N\) satisfies (31); \(\mathcal {R}_{\alpha }(\mathcal {X})\) is said to blow up stochastically for a given initial condition if \(\lim _{t \rightarrow \infty } \mathbb {E} X_i(t; \, \varvec{\alpha }) = \infty \) for some \(i \in \{1, 2, \ldots , N\}\), where the first-moment of the species copy-numbers \(\mathbb {E} \textbf{X}(t; \, \varvec{\alpha }) \in \mathbb {R}_{\ge }^N\) satisfies (33).
Nonnegative ODEs, such as Eqs. (31) and (33), need not have a nonnegative time-independent solution, i.e. the dynamics can be confined to an unbounded invariant set devoid of any equilibria. In this context, we introduce the following definition.
Definition A.4
(No-equilibrium catastrophes (NECs)) Reaction network \(\mathcal {R}_{\alpha }(\mathcal {X})\) is said to display a deterministic (respectively, a stochastic) no-equilibrium catastrophe (NEC) if, for all \(\varvec{\alpha } \in \mathbb {R}_{>}^a\) such that the RREs have no nonnegative equilibria, \(\mathcal {R}_{\alpha }(\mathcal {X})\) blows up deterministically (respectively, stochastically) for some nonnegative initial conditions.
Remark
More specifically, a nonnegative equilibrium can cease to exist by attaining a negative component, becoming complex, or vanishing all together. In this paper, we focus on the NEC that involves equilibria attaining negative components, which we call negative-equilibrium catastrophe and also abbreviate as NEC.
Appendix B: Stochastic biochemical control
In this section, we formulate the problem of achieving biochemical control over a given reaction network, starting with the following definition.
Definition B.1
(Black, grey and white box) Network \(\mathcal {R}_{\alpha }(\mathcal {X}) \ne \emptyset \) with unknown (respectively, only partially known) structure and dynamics is called a black-box (respectively, grey-box) network; \(\mathcal {R}_{\alpha }(\mathcal {X}) \ne \emptyset \) is called a white-box network if its structure and dynamics are completely known.
Given an input (uncontrolled) reaction network \(\mathcal {R}_{\alpha } = \mathcal {R}_{\alpha }(\mathcal {X})\), the objective of biochemical control is to design a controller network \(\mathcal {R}_{\beta ,\gamma }\) in order to ensure that the dynamics of desired input species \(\mathcal {X}\) is suitably controlled in the resulting output (controlled) network \(\mathcal {R}_{\alpha ,\beta ,\gamma } \equiv \mathcal {R}_{\alpha } \cup \mathcal {R}_{\beta ,\gamma }\); in what follows, we focus on black-box input networks. To this end, we partition the input species into \(\mathcal {X} = \mathcal {X}_{I} \cup \mathcal {X}_{\rho }\), where \(\mathcal {X}_{I}= \{X_1, X_2, \ldots , X_{N_{I}}\}\) are the \(1 \le N_{I} \le N\) interfacing species that can be interfaced with a given controller, while \(\mathcal {X}_{\rho } = \mathcal {X} {\setminus } \mathcal {X}_{I} = \{X_{N_{I}+1}, X_{N_{I}+2}, \ldots , X_N\} \) are the \(N_{\rho } = (N - N_{I})\) residual species that cannot be interfaced with the controller; input species whose dynamics we wish to control are called target species. The controller can be decomposed into two sub-networks, \(\mathcal {R}_{\beta ,\gamma } = \mathcal {R}_{\beta ,\gamma }(\mathcal {X}_{I},\mathcal {Y}) = \mathcal {R}_{\beta }(\mathcal {Y}) \cup \mathcal {R}_{\gamma }(\mathcal {X}_{I},\mathcal {Y})\), where \(\mathcal {R}_{\beta } = \mathcal {R}_{\beta }(\mathcal {Y})\), called the core, contains all the reactions that involve only the controlling species \(\mathcal {Y} = \{Y_1, Y_2, \ldots , Y_M\}\), while \(\mathcal {R}_{\gamma } = \mathcal {R}_{\gamma }(\mathcal {X}_{I},\mathcal {Y})\), called the interface, contains all of the remaining reactions, involving both \(\mathcal {X}_{I}\) and \(\mathcal {Y}\). Put more simply, \(\mathcal {R}_{\beta }\) describes internal dynamics of the controlling species, while \(\mathcal {R}_{\gamma }\) describes how the interfacing and controlling species interact.
In what follows, we focus on controlling average copy-number of a single target species which is interfacing; see [39] for a more general multi-species control of the full PMF when the targets are both interfacing and residual species. To this end, we denote the rate coefficients from the sub-networks \(\mathcal {R}_{\alpha }\), \(\mathcal {R}_{\beta }\) and \(\mathcal {R}_{\gamma }\) by \(\varvec{\alpha } \in \mathbb {R}_{>}^a\), \(\varvec{\beta } \in \mathbb {R}_{>}^b\) and \(\varvec{\gamma } \in \mathbb {R}_{>}^c\), respectively. We also let \(\mathbb {E} X_1(t; \, \cdot , \cdot \,; \, \varvec{\alpha }): \mathbb {R}_{>}^b \times \mathbb {R}_{>}^c \rightarrow \mathbb {R}\) be the first-moment of the target species \(X_1 \in \mathcal {X}_{I}\), which is a function of the control parameters \((\varvec{\beta },\varvec{\gamma })\) for every time t, input coefficient \(\varvec{\alpha }\) and every initial condition. More precisely, \(\mathbb {E} X_1 = \mathbb {E} X_1(t; \, \varvec{\beta }, \varvec{\gamma }; \, \varvec{\alpha }) = \langle x_1, p(\textbf{x}, \textbf{y}, t; \, \varvec{\beta },\varvec{\gamma }; \, \varvec{\alpha }) \rangle _{\textbf{x},\textbf{y}}\), where \(p(\textbf{x}, \textbf{y}, t; \, \varvec{\beta },\varvec{\gamma }; \, \varvec{\alpha })\) is the time-dependent PMF of the output network. In what follows, we denote the gradient operator with respect to \(\textbf{x}= (x_1, x_2, \ldots , x_N)\) by \(\varvec{\nabla }_{\textbf{x}} \equiv (\partial /\partial _{x_1}, \partial /\partial _{x_2}, \ldots , \partial /\partial _{x_N})\).
Definition B.2
(Control) Consider a black-box input network \(\mathcal {R}_{\alpha }(\mathcal {X})\), and the corresponding output network \(\mathcal {R}_{\alpha ,\beta ,\gamma }(\mathcal {X},\mathcal {Y}) = \mathcal {R}_{\alpha }(\mathcal {X}) \cup \mathcal {R}_{\beta ,\gamma }(\mathcal {X}_{I},\mathcal {Y})\). Assume we are given a nonempty open set \(\mathbb {S}_{\alpha } \subset \mathbb {R}_{>}^a\), stability index \(p \in \mathbb {Z}_{>}\), and a target value \(\bar{x}_1 \in \mathbb {R}_{>}\) together with a tolerance \(\varepsilon \in \mathbb {R}_{\ge }\). Then, the first-moment \(\mathbb {E} X_1 = \mathbb {E} X_1(t; \, \varvec{\beta }, \varvec{\gamma }; \, \varvec{\alpha })\) is said to be controlled in the long-run if there exists a nonempty open set \(\mathbb {S}_{\beta ,\gamma } \subset \mathbb {R}_{>}^{b+c}\) such that the following two conditions are satisfied for all initial conditions \((\textbf{X}(0), \textbf{Y}(0)) \in \mathbb {Z}_{\ge }^{N+M}\) and rate coefficients \((\varvec{\alpha }, \varvec{\beta }, \varvec{\gamma }) \in \mathbb {S}_{\alpha } \times \mathbb {S}_{\beta ,\gamma }\):
- (C.I):
-
Stability: \(\{\lim _{t \rightarrow \infty } \mathbb {E} X_i^p(t; \, \varvec{\beta }, \varvec{\gamma }; \, \varvec{\alpha }) < \infty \}_{i = 1}^N\) and \(\{\lim _{t \rightarrow \infty } \mathbb {E} Y_i^p(t; \, \varvec{\beta },\varvec{\gamma }; \, \varvec{\alpha }) < \infty \}_{i = 1}^M\).
- (C.II):
-
Accuracy: \(\lim _{t \rightarrow \infty } \Big |\mathbb {E} X_1(t; \, \varvec{\beta }, \varvec{\gamma }; \, \varvec{\alpha }) - \bar{x}_1 \Big | \le \varepsilon \), where \(\lim _{t \rightarrow \infty } \varvec{\nabla }_{\varvec{\beta }, \varvec{\gamma }} \mathbb {E} X_1(t; \, \varvec{\beta },\varvec{\gamma }; \, \varvec{\alpha }) \ne \textbf{0}\).
Condition (C.I) requires boundedness of the long-time moments, up to order \(p > 1\), of all of the species from the output network \(\mathcal {R}_{\alpha ,\beta ,\gamma }(\mathcal {X},\mathcal {Y})\). Minimally, one requires that the long-time first-moments are bounded, i.e. that the controller does not trigger a stochastic blow-up (see Definition A.3). The larger \(p \ge 1\) one chooses, the thinner the tail of the long-time output PMF, which ensures a more stable behavior of the output network. Condition (C.II) demands that the long-time average of \(X_1\), which is required to depend on at least one control parameter, is sufficiently close to the target value. Let us remark that (C.I)–(C.II) must hold within neighborhoods of the underlying rate coefficient values, reflecting the fact that measurement and fine-tuning of rate coefficients is not error-free. For the same reason, we have put forward a more relaxed accuracy criterion by allowing nonzero tolerance in condition (C.II).
1.1 Appendix B.1: Robust control
Condition (C.II) from Definition B.2 may be challenging to achieve due to the fact that the long-time first-moment \(\mathbb {E} X_1\) generally depends on the initial conditions, and on the input coefficients \(\varvec{\alpha }\), which motivates the following definition.
Definition B.3
(Robustness) Consider a black-box input network \(\mathcal {R}_{\alpha }(\mathcal {X})\), and the corresponding output network \(\mathcal {R}_{\alpha , \beta , \gamma }(\mathcal {X}, \mathcal {Y}) = \mathcal {R}_{\alpha }(\mathcal {X}) \cup \mathcal {R}_{\beta ,\gamma }(\mathcal {X}_{I},\mathcal {Y})\). Assume conditions (C.I)–(C.II) from Definition B.2 are satisfied. Then, network \(\mathcal {R}_{\beta ,\gamma }(\mathcal {X}_{I},\mathcal {Y})\) is a robust controller of the first-moment \(\mathbb {E} X_1\) in the long-run if the following two conditions are also satisfied:
- (R.I):
-
Robustness to initial conditions. There exists a unique stationary PMF \(p(\textbf{x}, \textbf{y}; \, \varvec{\beta },\varvec{\gamma }; \, \varvec{\alpha })\) such that \(\lim _{t \rightarrow \infty } \Vert p(\textbf{x}, \textbf{y}, t; \, \varvec{\beta },\varvec{\gamma }; \, \varvec{\alpha }) - p(\textbf{x}, \textbf{y}; \, \varvec{\beta },\varvec{\gamma }; \, \varvec{\alpha }) \Vert _{l_1} = 0\) for all \((\textbf{X}(0), \textbf{Y}(0)) \in \mathbb {Z}_{\ge }^{N+M}\) and \((\varvec{\alpha }, \varvec{\beta }, \varvec{\gamma }) \in \mathbb {S}_{\alpha } \times \mathbb {S}_{\beta ,\gamma }\).
- (R.II):
-
Robustness to input coefficients. The stationary first-moment, given by \(\mathbb {E} X_1^*(\varvec{\beta },\varvec{\gamma }; \, \varvec{\alpha })\) \(\equiv \langle x_1, p(\textbf{x}, \textbf{y}; \, \varvec{\beta },\varvec{\gamma }; \, \varvec{\alpha }) \rangle _{\textbf{x},\textbf{y}} > 0\), satisfies \(\varvec{\nabla }_{\varvec{\alpha }} \mathbb {E} X_1^*(\varvec{\beta },\varvec{\gamma }; \, \varvec{\alpha }) = \textbf{0}\) for all \((\varvec{\alpha }, \varvec{\beta }, \varvec{\gamma }) \in \mathbb {S}_{\alpha } \times \mathbb {S}_{\beta ,\gamma }\).
Remark
Robust controllers are also called integral-feedback controllers [19].
Remark
Conditions (C.II) and (R.II) demand that the first-moment of \(X_1\) is nondegenerate with respect to the control parameters, \(\varvec{\nabla }_{\varvec{\beta }, \varvec{\gamma }} \mathbb {E} X_1^*(\varvec{\beta },\varvec{\gamma }; \, \varvec{\alpha })\ne \textbf{0}\), and that it is degenerate with respect to the input parameters, \(\varvec{\nabla }_{\varvec{\alpha }} \mathbb {E} X_1^*(\varvec{\beta },\varvec{\gamma }; \, \varvec{\alpha }) = \textbf{0}\), respectively. As we prove in Lemma C.1 below, the degeneracy condition enforces singularity of an appropriate kinetic matrix.
Appendix C: Nonexistence of unimolecular integral-feedback controllers
Let \(\mathcal {R}_{\alpha }(\mathcal {X})\) be a black-box input network with a desired target species \(X_1 \in \mathcal {X}_{I}\), and let \(\mathcal {R}_{\beta ,\gamma }(\mathcal {X}_{I},\mathcal {Y}) = \mathcal {R}_{\beta }(\mathcal {Y}) \cup \mathcal {R}_{\gamma }(\mathcal {X}_{I},\mathcal {Y})\) be a unimolecular network. The first-moment equations for the output network \(\mathcal {R}_{\alpha , \beta , \gamma }(\mathcal {X}, \mathcal {Y}) = \mathcal {R}_{\alpha }(\mathcal {X}) \cup \mathcal {R}_{\beta ,\gamma }(\mathcal {X}_{I},\mathcal {Y})\) can be written in the following form:
where \(\mathcal {L}_{\alpha }\) is the (unknown) generator of the input network, \(\varvec{\beta }_{\cdot ,0} = (\beta _{1,0}, \beta _{2,0}, \ldots ,\beta _{M,0})\in \mathbb {R}_{\ge }^M\) is induced by the core network \(\mathcal {R}_{\beta }(\mathcal {Y})\), while cross-nonnegative matrix \(C^{1,1} \in \mathbb {R}^{N_{I} \times N_{I}}\) and nonnegative matrices \(C^{1,2} \in \mathbb {R}_{\ge }^{N_{I} \times M}\) and \(C^{2,1} \in \mathbb {R}_{\ge }^{M \times N_{I}}\) are induced by the interfacing network \(\mathcal {R}_{\gamma }(\mathcal {X}_{I},\mathcal {Y})\). Matrix \(\bar{C}^{2,2} \in \mathbb {R}_{\ge }^{M \times M}\) can be written as \(\bar{C}^{2,2} = (B + C^{2,2})\), with cross-nonnegative matrices B and \(C^{2,2}\) being induced by \(\mathcal {R}_{\beta }(\mathcal {Y})\) and \(\mathcal {R}_{\gamma }(\mathcal {X}_{I},\mathcal {Y})\), respectively. Note that \(\bar{C}^{2,2}\) encodes reactions that change the copy-numbers of the controlling species \(\mathcal {Y}\) either in \(\mathcal {X}_{I}\)-independent (via matrix B) or \(\mathcal {X}_{I}\)-dependent (via matrix \(C^{2,2}\)) manner. Let us also note that, by definition, \(\varvec{\beta } \in \mathbb {R}_{>}^b\) contains nonzero elements from vector \(\varvec{\beta }_{\cdot ,0}\), and absolute value of nonzero elements from matrix B; similarly, \(\varvec{\gamma } \in \mathbb {R}_{>}^c\) contains absolute values of nonzero elements from \(C^{1,1}\), \(C^{1,2}\), \(C^{2,1}\) and \(C^{2,2}\). We now present conditions on the matrices \(C^{2,1} = (\varvec{\gamma }_{\cdot ,1}^{2,1}, \varvec{\gamma }_{\cdot ,2}^{2,1}, \ldots , \varvec{\gamma }_{\cdot ,n}^{2,1})\) and \(\bar{C}^{2,2}\) that are necessary for unimolecular networks to exert robust (integral-feedback) control.
Lemma C.1
Assume a unimolecular reaction network \(\mathcal {R}_{\beta ,\gamma }(\mathcal {X}_{I},\mathcal {Y})\) is a robust controller for a black-box network \(\mathcal {R}_{\alpha }(\mathcal {X})\). Then, \(\bar{C}^{2,2}\) from (34) is a singular matrix all eigenvalues of which have nonpositive real parts for all \((\varvec{\beta }, \varvec{\gamma }) \in \mathbb {S}_{\beta ,\gamma }\). Furthermore, the first column of matrix \(C^{2,1}\) is nonzero.
Proof
Assume \(\bar{C}^{2,2}\) has an eigenvalue with a positive real part for some \((\varvec{\beta }, \varvec{\gamma }) \in \mathbb {S}_{\beta ,\gamma }\); it then follows from (34) that \(\lim _{t \rightarrow \infty } \mathbb {E} Y_i = \infty \) for some \(i \in \{1, 2, \ldots , M\}\). Therefore, condition (C.I) from Definition B.2 does not hold for all \((\varvec{\beta }, \varvec{\gamma }) \in \mathbb {S}_{\beta ,\gamma }\).
Assume \(\bar{C}^{2,2}\) is nonsingular for some \((\varvec{\beta }, \varvec{\gamma }) \in \mathbb {S}_{\beta ,\gamma }\). Let \((\mathbb {E} \textbf{X}_{I}^*, \mathbb {E} \textbf{X}_{\rho }^*, \mathbb {E} \textbf{Y}^*)\) be the unique stationary first-moments, obtained by setting to zero the left-hand side in (34), and in the ODEs for higher-order moments. Then, one can eliminate \(\mathbb {E} \textbf{Y}^*\) via \(\mathbb {E} \textbf{Y}^* = - (\bar{C}^{2,2})^{-1} (\varvec{\beta }_{\cdot ,0} + C^{2,1} \mathbb {E} \textbf{X}_{I}^*)\). The remaining system of equations for \((\mathbb {E} \textbf{X}_{I}^*, \mathbb {E} \textbf{X}_{\rho }^*)\), and hence \(\mathbb {E} X_1^*\), depends on \(\varvec{\alpha }\) via the unknown generator \(\mathcal {L}_{\alpha }\). Therefore, condition (R.II) from Definition B.3 does not hold for all \((\varvec{\beta }, \varvec{\gamma }) \in \mathbb {S}_{\beta ,\gamma }\).
Assume the first column of \(C^{2,1}\) is zero, \(\varvec{\gamma }_{\cdot ,1}^{2,1} = \textbf{0}\). By the Fredholm alternative theorem, a necessary condition for the stationary first-moment \(\mathbb {E} \textbf{Y}^*\) to exist is then given by \((\langle \textbf{w}, \varvec{\beta }_{\cdot ,0} \rangle + \sum _{i = 2}^n \langle \textbf{w}, \varvec{\gamma }_{\cdot ,i}^{2,1}\rangle \mathbb {E} X_i^*) = 0\) for every \(\textbf{w} \in \mathbb {R}^M\) such that \((\bar{C}^{2,2})^{\top } \textbf{w} = \textbf{0}\). The stationary average \(\mathbb {E} X_1^*\) is not uniquely determined by these constraints and, hence, depends on \(\varvec{\alpha }\). Therefore, condition (R.II) from Definition B.3 does not hold. \(\square \)
Theorem C.1
There does not exist a unimolecular integral-feedback controller.
Proof
Suppose, for contradiction, that \(\mathcal {R}_{\beta ,\gamma }(\mathcal {X}_{I},\mathcal {Y})\) is a robust (integral-feedback) controller, i.e. that the corresponding output network \(\mathcal {R}_{\alpha , \beta , \gamma }(\mathcal {X}, \mathcal {Y}) = \mathcal {R}_{\alpha }(\mathcal {X}) \cup \mathcal {R}_{\beta ,\gamma }(\mathcal {X}_{I},\mathcal {Y})\) satisfies Definition B.3. Lemma (C.1) then implies that \((\bar{C}^{2,2})^{\top }\) is a singular cross-nonnegative matrix all eigenvalues of which have nonpositive real parts; in what follows, we assume that \((\bar{C}^{2,2})^{\top }\) is irreducible. It then follows from [35, Theorem 5.6] that there exists a positive vector \(\textbf{w} \in \mathbb {R}_{>}^N\) such that \((\bar{C}^{2,2})^{\top } \textbf{w} = \textbf{0}\), so that \(\textrm{d}/\textrm{d} t \langle \textbf{w}, \mathbb {E} \textbf{Y} \rangle = (\langle \textbf{w}, \varvec{\beta }_{\cdot ,0} \rangle + \sum _{i = 1}^n \langle \textbf{w}, \varvec{\gamma }_{\cdot ,i}^{2,1}\rangle \mathbb {E} X_i)\). Using the fact that, by Lemma (C.1), \(\varvec{\gamma }_{\cdot ,1}^{2,1} \ne \textbf{0}\), and that, by Definition B.3, \(\lim _{t \rightarrow \infty } \mathbb {E} X_1(t; \, \varvec{\beta }, \varvec{\gamma }; \, \varvec{\alpha }) = \mathbb {E} X_1^*(\varvec{\beta },\varvec{\gamma }) > 0\), it follows that \(\textrm{d}/\textrm{d} t \langle \textbf{w}, \mathbb {E} \textbf{Y} \rangle > 0\) in the long-run. Therefore, \(\lim _{t \rightarrow \infty } \mathbb {E} Y_i = \infty \) for some \(i \in \{1, 2, \ldots , M\}\), implying that condition (C.I) from Definition B.2 does not hold. Hence, \(\mathcal {R}_{\beta ,\gamma }(\mathcal {X}_{I},\mathcal {Y})\) is not a robust controller. If \(\bar{C}^{2,2}\) is reducible, then analogous argument can be applied to each of the underlying irreducible components, implying the statement of the theorem. Due to linearity of the controller, the same argument implies that a deterministic integral-feedback controller does not exist. \(\square \)
The proof of Theorem C.1 shows that a unimolecular integral-feedback controller does not exist because the underlying reaction network \(\mathcal {R}_{\beta ,\gamma }(\mathcal {X}_{I},\mathcal {Y})\) triggers a deterministic and stochastic NEC (see also Definition A.4 in Sect. A.2.3) for all nonnegative initial conditions, violating the stability condition (C.I) from Definition B.2. For example, taking \(C^{2,1} = (\varvec{\gamma }_{\cdot ,1}^{2,1}, \textbf{0}, \ldots , \textbf{0})\), it follows that the deterministic equilibrium of the target species is \(x_1^* = -\langle \textbf{w}, \varvec{\beta }_{\cdot ,0} \rangle /\langle \textbf{w},\varvec{\gamma }_{\cdot ,1}^{2,1} \rangle < 0\).
Appendix D: Hyperbolic kinetic transformation
In this section, we briefly discuss how to map arbitrary polynomial ODEs into dynamically similar mass-action RREs [20]. To this end, let us note that every component of every polynomial function \(\varvec{\mathcal {P}}(\textbf{x}; \, \varvec{\alpha }) = (\mathcal {P}_1(\textbf{x}; \, \varvec{\alpha }), \mathcal {P}_2(\textbf{x}; \, \varvec{\alpha }),\) \( \ldots , \mathcal {P}_N(\textbf{x}; \, \varvec{\alpha }))^{\top } \in \mathbb {P}_{m}(\mathbb {R}^N; \, \mathbb {R}^N)\), where \(\varvec{\alpha } \in \mathbb {R}_{>}^a\), can be written as
Here, \(\mathcal {A}_{i}^{+}\) and \(\mathcal {A}_{i}^{-} \equiv (\mathcal {A}_{i, \mathcal {K}}^{-} \cup \mathcal {A}_{i, \mathcal {N}}^{-})\) are the indices of all of the distinct positive and negative terms in \(\mathcal {P}_i(\textbf{x}; \,\varvec{\alpha })\), respectively, where \(\mathcal {A}_{i, \mathcal {K}}^{-} = \{j \in \mathcal {A}_i^{-} | \nu _{i,j} \ne 0\}\) and \(\mathcal {A}_{i, \mathcal {N}}^{-} = \{j \in \mathcal {A}_i^{-} | \nu _{i,j} = 0\}\).
Definition D.1
(Cross-negative terms; kinetic functions) Consider a polynomial function \(\varvec{\mathcal {P}}(\textbf{x}; \, \varvec{\alpha }) \in \mathbb {P}_{m}(\mathbb {R}^N; \, \mathbb {R}^N)\) with \(\varvec{\alpha } \in \mathbb {R}_{>}^a\), whose i-component is given by (35). Monomials \(\{- \alpha _j \textbf{x}^{\varvec{\nu }_{j, \cdot }} \}_{j \in \mathcal {A}_{i, \mathcal {N}}^{-}}\) are called cross-negative terms. Polynomial functions without cross-negative terms, \(\mathcal {A}_{i, \mathcal {N}}^{-} = \emptyset \), are called kinetic functions and denoted by \(\varvec{\mathcal {K}}(\textbf{x}; \, \varvec{\alpha })\); the set of all kinetic functions of degree at most m is denoted by \(\mathbb {P}_{m}^{\mathcal {K}}(\mathbb {R}^N; \, \mathbb {R}^N)\). Polynomial functions with a cross-negative term, \(\mathcal {A}_{i, \mathcal {N}}^{-} \ne \emptyset \), are called non-kinetic functions and denoted by \(\varvec{\mathcal {N}}(\textbf{x}; \, \varvec{\alpha })\); the set of all non-kinetic functions of degree at most m is denoted by \(\mathbb {P}_{m}^{\mathcal {N}}(\mathbb {R}^N; \, \mathbb {R}^N)\).
Remark
Definition D.1 captures the fact that biochemical reactions cannot consume a species when the concentration of that species is zero, e.g. see network (1); Theorem A.1 is a direct consequence of the absence of cross-negative terms in kinetic functions [14].
In order to map an arbitrary input non-kinetic function \(\varvec{\mathcal {N}}(\textbf{x}; \, \varvec{\alpha }) \in \mathbb {P}_{m}^{\mathcal {N}}(\mathbb {R}^N; \, \mathbb {R}^N)\) into an output kinetic one \(\varvec{\mathcal {K}}(\bar{\textbf{x}}; \, \bar{\varvec{\alpha }}) \in \mathbb {P}_{\bar{m}}^{\mathcal {K}}(\mathbb {R}^{\bar{N}}; \, \mathbb {R}^{\bar{N}})\), while preserving desired dynamical features over suitable time-intervals, a number of so-called kinetic transformations have been developed in [20]. Such mappings involve a dimension expansion (i.e. an introduction of additional biochemical species, \(\bar{N} \ge N\)) or an increase in non-linearity (\(\bar{m} \ge m\)). We now present one such kinetic transformation.
Definition D.2
(Hyberbolic transformation) Consider a system of polynomial ODEs given by
with the index sets as defined in (35). Consider also the RREs given by
with a parameter \(\varepsilon \in \mathbb {R}_{>}\). Kinetic transformation \(\Psi _{H}^{\varepsilon }: \mathbb {P}_{m}(\mathbb {R}^N; \, \mathbb {R}^N) \rightarrow \mathbb {P}_{\bar{m}}^{\mathcal {K}}(\mathbb {R}^{(2 N - n)}; \, \mathbb {R}^{(2 N -n)})\), mapping the right-hand side of ODEs (36) to the right-hand side of (37), with \(\bar{m} \le m + 2\), is called a hyberbolic transformation.
By comparing equilibria of (36) and (37), the following proposition is established.
Proposition D.1
Let \(\textbf{x}^* = (x_1^*, x_2^*, \ldots , x_N^*) \in \mathbb {R}^N\) be an equilibrium of the ODE system (36) with \(x_{i}^* \ne 0\) for all \(i \in \{n+1, n+2, \ldots , N\}\). Then, \((\textbf{x}^*, \bar{\textbf{x}}^*) \in \mathbb {R}^{2 N - n}\) is an equilibrium of the ODE system (37) for some \(\bar{\textbf{x}}^* \in \mathbb {R}^{N - n}\).
Let us note that, while the \(\textbf{x}\)-equilibria are preserved by the hyperbolic kinetic transformation, their properties, such as stability, are not necessarily preserved; a stronger dynamical preservation is ensured by taking \(\varepsilon \) sufficiently small.
Theorem D.1
Solutions of (36), with \((x_{n+1}, x_{n+2}, \ldots , x_{N}) \in \mathbb {R}_{>}^{N-n}\), are asymptotically equivalent to the solutions of (37) in the limit \(\varepsilon \rightarrow 0\).
Proof
Let us introduce a change of coordinates \(y_i = (x_i - \bar{x}_i)\) for \(i \in \{n+1, n+2, \ldots , N\}\), leading to a regular singularly perturbed system in the variables \((x_1, x_2, \ldots , x_n, y_{n+1}, y_{n+2}, \ldots , y_{N},\bar{x}_{n+1}, \bar{x}_{n+2},\) \(\ldots ,\bar{x}_N)\). The adjoined sub-system has an isolated equilibrium \((\bar{x}_{n+1}^*, \bar{x}_{n+2}^*, \ldots , \bar{x}_N^*) = \textbf{0}\) which, when substituted into the degenerate sub-system in the variables \((x_1, x_2, \ldots , x_n,\) \( y_{n+1}, y_{n+2},\) \(\ldots ,y_{N})\), leads to (36). If \((x_{n+1}, x_{n+2}, \ldots , x_{N})\) \(\in \mathbb {R}_{>}^{N-n}\), then the trivial adjoined equilibrium is stable, and Tikhonov’s theorem [40] implies the statement of Theorem D.1. \(\square \)
Appendix E: Robust control of unimolecular input networks
In this section, we analyze performance of the class of second-order integral-feedback controllers \(\mathcal {R}_{\beta ,\gamma }(\{X_1, X_2\}, \{Y_1, Y_2\}) = \mathcal {R}_{\beta }(Y_1, Y_2) \cup \mathcal {R}_{\gamma }^{0}(Y_2; \, X_1) \cup \mathcal {R}_{\gamma }^{+}(X_i; \, Y_1) \cup \mathcal {R}_{\gamma }^{-}(X_j; \, Y_2)\), given by (8), by embedding them into the class of unimolecular input networks whose RREs, given by
have an asymptotically stable equilibrium. We also assume the input network has two interfacing species \(X_{I} = \{X_1, X_2\}\), and the goal is to control the concentration/first-moment of the target species \(X_1\).
In what follows, we let \(A_{(i_1, i_2, \ldots , i_n),(j_1, j_2, \ldots , j_m)} \in \mathbb {R}^{(N-n) \times (N-m)}\) denote the sub-matrix obtained by removing from \(A \in \mathbb {R}^{N \times N}\) the rows \(\{i_1, i_2, \ldots , i_n\} \subset \{1, 2\, \ldots , N\}\) and columns \(\{j_1, j_2, \ldots , j_m\}\) \(\subset \{1, 2\, \ldots , N\}\). Furthermore, we let \(A_{\varvec{\alpha }_{\cdot , j} \rightarrow \textbf{x}} \in \mathbb {R}^{N \times N}\) denote the matrix obtained by replacing the j-column of A by a vector \(\textbf{x} \in \mathbb {R}^N\).
Theorem E.1
(Deterministic equilibria) Let \(\mathcal {R}_{\alpha } = \mathcal {R}_{\alpha }(\mathcal {X})\), with species \(\mathcal {X} = \{X_1, X_2, \ldots , X_N\}\), be the class of unimolecular input networks whose RREs (38) have an asymptotically stable equilibrium. Let \(\mathcal {R}_{\beta ,\gamma } = \mathcal {R}_{\beta ,\gamma }(\{X_1, X_2\}, \{Y_1, Y_2\})\) be the controller given by (8). Then, the output network \(\mathcal {R}_{\alpha ,\beta ,\gamma } = \mathcal {R}_{\alpha } \cup \mathcal {R}_{\beta ,\gamma }\) satisfies the following properties:
- (i):
-
Pure positive interfacing. Let \(\mathcal {R}_{\gamma }^{-} = \emptyset \). If positive interfacing is direct, \(\mathcal {R}_{\gamma }^{+} = \mathcal {R}_{\gamma }^{+}(X_1, Y_1) \ne \emptyset \), then there exists a nonnegative equilibrium if and only if \(\frac{\beta _0}{\gamma _1} > \frac{|A_{\varvec{\alpha }_{\cdot ,1} \rightarrow -\varvec{\alpha }_{\cdot ,0}}|}{|A|}\); the same is true if positive interfacing is indirect, \(\mathcal {R}_{\gamma }^{+} = \mathcal {R}_{\gamma }^{+}(X_2, Y_1) \ne \emptyset \), and if \(|A_{2,1}| \ne 0\).
- (ii):
-
Pure negative interfacing. Let \(\mathcal {R}_{\gamma }^{+} = \emptyset \). If negative interfacing is direct, \(\mathcal {R}_{\gamma }^{-} = \mathcal {R}_{\gamma }^{-}(X_1, Y_2) \ne \emptyset \), then there exists a nonnegative equilibrium if and only if \(\frac{\beta _0}{\gamma _1} < \frac{|A_{1 \rightarrow -\varvec{\alpha }_{\cdot ,0}}|}{|A|}\). If negative interfacing is indirect, \(\mathcal {R}_{\gamma }^{-} = \mathcal {R}_{\gamma }^{-}(X_2, Y_2) \ne \emptyset \), and if \(|A_{2,1}| \ne 0\), then there exists a nonnegative equilibrium if and only if \(\frac{|(A_{\varvec{\alpha }_{\cdot ,1} \rightarrow -\varvec{\alpha }_{\cdot ,0}})_{2,2}|}{|A_{2,2}|}< \frac{\beta _0}{\gamma _1} < \frac{|A_{\varvec{\alpha }_{\cdot ,1} \rightarrow -\varvec{\alpha }_{\cdot ,0}}|}{|A|}\).
- (iii):
-
Combined indirect negative interfacing. Let \(\mathcal {R}_{\gamma }^{-} = \mathcal {R}_{\gamma }^{-}(X_2, Y_2) \ne \emptyset \). If positive interfacing is direct, \(\mathcal {R}_{\gamma }^{+} = \mathcal {R}_{\gamma }^{+}(X_1, Y_1) \ne \emptyset \), then there exists a nonnegative equilibrium if and only if \(\frac{\beta _0}{\gamma _1} > \frac{|(A_{\varvec{\alpha }_{\cdot ,1} \rightarrow -\varvec{\alpha }_{\cdot ,0}})_{2,2}|}{|A_{2,2}|}\); the same is true if positive interfacing is indirect, \(\mathcal {R}_{\gamma }^{+} = \mathcal {R}_{\gamma }^{+}(X_2, Y_1) \ne \emptyset \), and if \(|A_{2,1}| \ne 0\).
- (iv):
-
Combined direct negative interfacing. Let \(\mathcal {R}_{\gamma }^{-} = \mathcal {R}_{\gamma }^{-}(X_1, Y_2) \ne \emptyset \). If positive interfacing is direct, \(\mathcal {R}_{\gamma }^{+} = \mathcal {R}_{\gamma }^{+}(X_1, Y_1) \ne \emptyset \), then there exists a nonnegative equilibrium for any choice of \(\varvec{\beta } \in \mathbb {R}_{>}^{2}\) and \(\varvec{\gamma } \in \mathbb {R}_{>}^3\); the same is true if positive interfacing is indirect, \(\mathcal {R}_{\gamma }^{+} = \mathcal {R}_{\gamma }^{+}(X_2, Y_1) \ne \emptyset \), and if \(|A_{2,1}| \ne 0\).
Proof
Equilibria of the output network, denoted by \((\textbf{x}^*, y_1^*, y_2^*) \in \mathbb {R}^{N+2}\), satisfy
where \((\varvec{\alpha }_{\cdot , 0} + A \textbf{x})\) is the unknown kinetic function from (38), see also Lemma A.1. Statements (iii) and (iv) of the theorem are now proved; statements (i) and (ii) follow analogously.
Case \(\mathcal {R}_{\gamma }^{+} = \mathcal {R}_{\gamma }^{+}(X_1, Y_1) \ne \emptyset \) and \(\mathcal {R}_{\gamma }^{-} = \mathcal {R}_{\gamma }^{-}(X_2, Y_2) \ne \emptyset \). Defining vectors \(\bar{\textbf{x}}^* \equiv (x_2^*, x_3^*, \ldots , x_N^*)^{\top }\), \({\bar{\varvec{\alpha }}}_{\cdot , 0} \equiv (\alpha _{2,0}, \alpha _{3,0}, \ldots , \alpha _{N,0})^{\top }\), \({\bar{\varvec{\alpha }}}_{1, \cdot } \equiv (\alpha _{1,2}, \alpha _{1,3}, \ldots , \alpha _{1,N})^{\top }\) and \({\bar{\varvec{\alpha }}}_{\cdot , 1} \equiv (\alpha _{2,1}, \alpha _{3,1}, \ldots , \alpha _{N,1})^{\top }\), Eq. (39) can be written as
Assume there exists \(y_2^* > 0\). Since all eigenvalues of A have negative real parts, the same is true for \(A_{1,1}\), implying nonpositivity of matrix \(\left( A_{1,1} - \gamma _{3} y_2^* \, \textrm{diag}(\textbf{e}_1) \right) ^{-1} \in \mathbb {R}_{\le }^{(N-1) \times (N-1)}\) [35, Theorem 4.3], so that \(\bar{\textbf{x}}^* \in \mathbb {R}_{\ge }^{N-1}\). Matrix \(\left( A_{1,1} - \gamma _{3} y_2^* \, \textrm{diag}(\textbf{e}_1) \right) ^{-1}\) can be written in a cofactor form
where \(C_{i.j}\) and \(D_{i,j}\) are the (i, j)-cofactors of \(A_{1,1}\) and \(A_{(1,2), (1,2)}\), respectively. Substituting (41) into (40), one obtains the quadratic equation
Since A is cross-nonnegative with a negative spectral abscissa, it follows that \(|A_{2,2}|/|A_{1,1}| > 0\), \(|A|/|A_{1,1}| < 0\), \(|A_{(1,2),(1,2)}|/|A_{1,1}| < 0\), \(|(A_{\varvec{\alpha }_{\cdot ,1} \rightarrow -\varvec{\alpha }_{\cdot ,0}})_{2,2}|/|A_{2,2}| \ge 0\), and \(|A_{\varvec{\alpha }_{\cdot ,1} \rightarrow -\varvec{\alpha }_{\cdot ,0}}|/|A| \ge 0\). If \((\beta _0/\gamma _1 - |(A_{\varvec{\alpha }_{\cdot ,1} \rightarrow -\varvec{\alpha }_{\cdot ,0}})_{2,2}|/|A_{2,2}|) > 0\), it follows from (42) that there exists an equilibrium \((y_1^*, y_2^*) \in \mathbb {R}_{>}^{2}\), consistent with the assumption. On the other hand, if \((\beta _0/\gamma _1 - |(A_{\varvec{\alpha }_{\cdot ,1} \rightarrow -\varvec{\alpha }_{\cdot ,0}})_{2,2}|/|A_{2,2}|) \le 0\), using the fact that \(|A_{\varvec{\alpha }_{\cdot ,1} \rightarrow -\varvec{\alpha }_{\cdot ,0}}|/|A| \ge |(A_{\varvec{\alpha }_{\cdot ,1} \rightarrow -\varvec{\alpha }_{\cdot ,0}})_{2,2}|/|A_{2,2}|\), it follows that \((y_1^*, y_2^*) \notin \mathbb {R}_{>}^{2}\).
Case \(\mathcal {R}_{\gamma }^{+} = \mathcal {R}_{\gamma }^{+}(X_2, Y_1) \ne \emptyset \) and \(\mathcal {R}_{\gamma }^{-} = \mathcal {R}_{\gamma }^{-}(X_2, Y_2) \ne \emptyset \). Equation (39) reads
from which, using (41), one obtains
If \(|A_{2,1}| \ne 0\) and \((\beta _0/\gamma _1 - |(A_{\varvec{\alpha }_{\cdot ,1} \rightarrow - \varvec{\alpha }_{\cdot ,0}})_{2,2}|/|A_{2,2}|) > 0\), using the fact that \(|A_{2,1}|/|A_{1,1}| < 0\), it follows that there exists an equilibrium \((y_1^*, y_2^*) \in \mathbb {R}_{>}^{2}\). If \(|A_{2,1}| \ne 0\) and \((\beta _0/\gamma _1 - |(A_{\varvec{\alpha }_{\cdot ,1} \rightarrow - \varvec{\alpha }_{\cdot ,0}})_{2,2}|/|A_{2,2}|) \le 0\), then \((y_1^*, y_2^*) \notin \mathbb {R}_{>}^{2}\).
Case \(\mathcal {R}_{\gamma }^{+} = \mathcal {R}_{\gamma }^{+}(X_1, Y_1) \ne \emptyset \) and \(\mathcal {R}_{\gamma }^{-} = \mathcal {R}_{\gamma }^{-}(X_1, Y_2) \ne \emptyset \). Equation (39) reads
from which it follows that there exists \((y_1^*, y_2^*) \in \mathbb {R}_{>}^{2}\) for any choice of \(\varvec{\beta } \in \mathbb {R}_{>}^{2}\) and \(\varvec{\gamma } \in \mathbb {R}_{>}^3\).
Case \(\mathcal {R}_{\gamma }^{+} = \mathcal {R}_{\gamma }^{+}(X_2, Y_1) \ne \emptyset \) and \(\mathcal {R}_{\gamma }^{-} = \mathcal {R}_{\gamma }^{-}(X_1, Y_2) \ne \emptyset \). Equation (39) reads
Assume there exists \(y_1^* > 0\). Then, it follows that \(\bar{\textbf{x}}^* \in \mathbb {R}_{\ge }^{N-1}\). Substituting (41) with \(\gamma _3 = 0\) into (46), one obtains the quadratic equation
If \(|A_{2,1}| \ne 0\), there exists an equilibrium \((y_1^*, y_2^*) \in \mathbb {R}_{>}^{2}\) for any choice of \(\varvec{\beta } \in \mathbb {R}_{>}^{2}\) and \(\varvec{\gamma } \in \mathbb {R}_{>}^3\), consistent with the assumption. \(\square \)
Remark
We say that an input network is degenerate if its \(x_1\)-component of the asymptotically stable equilibrium satisfies \(x_1^{**} = |A_{\varvec{\alpha }_{\cdot ,1} \rightarrow -\varvec{\alpha }_{\cdot ,0}}|/|A| = 0\) or \((x_1^{**})|_2 = |(A_{\varvec{\alpha }_{\cdot ,1} \rightarrow -\varvec{\alpha }_{\cdot ,0}})_{2,2}|/|A_{2,2}| = 0\); otherwise, the network is said to be nondegenerate. If \(x_1^{**} = 0\), then, in addition to the case (iv), cases (i) and (iii) from Theorem E.1 also generically ensure existence of a nonnegative equilibrium; case (ii) then necessarily leads to negative equilibria. For example, a degenerate network is obtained by neglecting basal production (setting \(\alpha _0 = 0\)) in (9).
Let us note that Theorem E.1 has an intuitive interpretation. In particular, \(x_1^{**} = |A_{\varvec{\alpha }_{\cdot ,1} \rightarrow -\varvec{\alpha }_{\cdot ,0}}|/|A|\) is the \(x_1\)-component of the equilibrium of the input network, \((x_1^{**})|_2 = |(A_{\varvec{\alpha }_{\cdot ,1} \rightarrow -\varvec{\alpha }_{\cdot ,0}})_{2,2}|/|A_{2,2}|\) is the \(x_1\)-component of the equilibrium of the restricted input network \(\mathcal {R}_{\alpha }(\mathcal {X} \setminus X_2)\) with species concentration \(x_2 \equiv 0\). Furthermore, condition \(|A_{2,1}| \ne 0\) requires that the species \(X_2\) influences \(X_1\) in the input network; more precisely, \(|A_{2,1}| \ne 0\) requires that there exists at least one directed path from \(X_2\) to \(X_1\) in the digraph induced by the Jacobian matrix of \(\mathcal {R}_{\alpha }(\mathcal {X})\).
We now prove that if, for a particular choice of the rate coefficients, the output network from Theorem E.1 has no nonnegative equilibria, then the network displays deterministic and stochastic blow-ups, i.e. we prove that the output network undergoes deterministic and stochastic NECs (see also Definition A.4 in Sect. 1). In what follows, the critical values of \(\beta _0/\gamma _1\) at which nonnegative equilibria cease to exist in Theorem E.1 are called bifurcation points.
Theorem E.2
(Negative-equilibrium catastrophe) Consider a unimolecular input network \(\mathcal {R}_{\alpha }\) whose RREs have an asymptotically stable equilibrium, and a controller \(\mathcal {R}_{\beta ,\gamma }\) of the form (8). Then, excluding the bifurcation points, the output network \(\mathcal {R}_{\alpha ,\beta ,\gamma } = \mathcal {R}_{\alpha } \cup \mathcal {R}_{\beta ,\gamma }\) displays deterministic and stochastic negative-equilibrium catastrophe for all nonnegative initial conditions.
Proof
Consider the case with pure direct positive interfacing from Theorem E.1(i); the RREs for the concentration \(\textbf{x} = (x_1, x_2, \ldots , x_N) \in \mathbb {R}_{\ge }^N\) and \((y_2 - y_1) \in \mathbb {R}^2\) are given by
Let \(\textbf{w} \equiv |A|^{-1} (C_{1,1}, C_{2,1}, \ldots , C_{N,1}) \in \mathbb {R}^{N}\), where \(C_{i,j}\) is the (i, j)-cofactor of matrix \(A \in \mathbb {R}^{N \times N}\). Taking the inner product \(\langle \textbf{w}, \cdot \rangle \) in the first equation from (48), and using the fact that \(\langle A^{\top } \textbf{w}, \textbf{x} \rangle = x_1\), one obtains:
Using the fact that \(|A_{1,1}|/|A| < 0\), Eqs. (48) and (49) imply that
By Theorem E.1(i), a nonnegative equilibrium does not exist if and only if \(\frac{\beta _0}{\gamma _1} \le \frac{|A_{\varvec{\alpha }_{\cdot ,1} \rightarrow -\varvec{\alpha }_{\cdot ,0}}|}{|A|}\); excluding the bifurcation point \(\frac{\beta _0}{\gamma _1} = \frac{|A_{\varvec{\alpha }_{\cdot ,1} \rightarrow -\varvec{\alpha }_{\cdot ,0}}|}{|A|}\), it follows from (50) that the linear combination \((-\langle \textbf{w}, \textbf{x} \rangle + \gamma _{1}^{-1} \left( y_2 - y_1\right) )\), and hence an underlying concentration, is a monotonically increasing function of time for all nonnegative initial conditions, i.e. the output network displays a deterministic NEC. Identical argument implies that then the output network displays a stochastic NEC as well. Deterministic and stochastic NECs for cases (ii) and (iii) from Theorem E.1 are established analogously. \(\square \)
Appendix F: Robust control of higher-molecular input networks
Consider an arbitrary (unimolecular, bimolecular or any higher-molecular) mass-action input network \(\mathcal {R}_{\alpha } = \mathcal {R}_{\alpha }(\mathcal {X})\), whose RREs are given by
Furthermore, let \(\mathcal {R}_{\beta ,\gamma }^{\pm }(X_i, Y_{1,i}, Y_{2,i}) \equiv \Big (\mathcal {R}_{\beta }(Y_{1,i}, Y_{2,i}) \cup \mathcal {R}_{\gamma }^0(Y_{2,i}; \, X_i) \cup \mathcal {R}_{\gamma }^{+}(X_{i}; \, Y_{1,i}) \cup \mathcal {R}_{\gamma }^{-}(X_{i}; \, Y_{2,i}) \Big )\) be the controller given by
In what follows, we let \(\textbf{y} = (y_{1,1}, y_{2,1}, y_{1,2}, y_{2,2}, \ldots , y_{1,N}, y_{2,N}) \in \mathbb {R}_{\ge }^{2 N}\) denote the concentration vector for the species \(\mathcal {Y} = \{Y_{1,1}, Y_{2,1}, Y_{1,2}, Y_{2,2}, \ldots , Y_{1,N}, Y_{2,N} \}\).
Theorem F.1
(Positive equilibrium) Let \(\mathcal {R}_{\alpha } = \mathcal {R}_{\alpha }(\mathcal {X})\), with species \(\mathcal {X} = \{X_1, X_2, \ldots , X_N\}\), be an arbitrary mass-action input network with the RREs (51). Let \(\bigcup _{i = 1}^N \mathcal {R}_{\beta ,\gamma }^{\pm }(X_i, Y_{1,i}, Y_{2,i})\) be the controller with \(\mathcal {R}_{\beta ,\gamma }^{\pm }(X_i, Y_{1,i}, Y_{2,i})\) given by (52). Then, for any choice of the rate coefficients \(\varvec{\alpha }\), \(\varvec{\beta }\) and \(\varvec{\gamma }\), the output network \(\mathcal {R}_{\alpha ,\beta ,\gamma }(\mathcal {X}, \mathcal {Y}) = \mathcal {R}_{\alpha }(\mathcal {X}) \bigcup _{i = 1}^N \mathcal {R}_{\beta ,\gamma }^{\pm }(X_i, Y_{1,i}, Y_{2,i})\) has a positive equilibrium \((\textbf{x}^*, \textbf{y}^*) \in \mathbb {R}_{>}^{3 N}\), with \(x_i^* = (\beta _{0,i}/\gamma _{1,i}) > 0\) for all \(i \in \{1, 2, \ldots , N\}\).
Proof
The RREs of the output network \(\mathcal {R}_{\alpha }(\mathcal {X}) \bigcup _{i = 1}^N \mathcal {R}_{\beta ,\gamma }^{\pm }(X_i, Y_{1,i}, Y_{2,i})\) are given by
and the statement of the theorem follows using the same argument as in (7) from Sect. 3. \(\square \)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Plesa, T., Dack, A. & Ouldridge, T.E. Integral feedback in synthetic biology: negative-equilibrium catastrophe. J Math Chem 61, 1980–2018 (2023). https://doi.org/10.1007/s10910-023-01495-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10910-023-01495-3