Abstract
In recent years, hybrid nanoflowers (hNFs), the newest class of nanoparticles, have been highly preferred due to their excellent activity and stability. In this study, hybrid nanoflower synthesis was carried out using N1-phenylbenzene-1,2-diamine and 1,2-phenylenediamine as the organic part and copper(II) metal ions as the inorganic part. In the first stage, the characterization of the synthesized hybrid nanoflowers was carried out using various techniques. For the characterization of the synthesized hNFs, structure elucidation was performed using Scanning Electron Microscopy (SEM), Energy Dispersive X-ray spectroscopy (EDX), Fourier transform infrared spectrometry (FT-IR), X-ray diffraction (XRD) spectroscopy and elemental mapping. In the other study stage, the cytotoxic effects of hybrid nanoflowers were evaluated using A549 and MCF7 cell lines. When 1,2-phenylenediamine and N1-phenylbenzene-1,2-diamine were converted into CuhNFs, it was effective in MCF7 and A549 cell lines. Docking studies were performed using the Prime MM-GBSA method to estimate binding affinities and determine the binding mode. ADME analysis was performed using the Schrödinger 2021-2 QikProp wizard. Support was obtained from molecular docking to confirm the potential of N1-phenylbenzene-1,2-diamine and 1,2-phenylenediamine compounds for both breast and lung cancer. Molecular docking studies can provide information about binding interactions between compounds with identified targets, which may explain their inhibitory activity. A better result can be obtained by examining the binding patterns in the active binding region of the compounds through molecular docking.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Nanotechnology is promising in fields like biocatalysis and the development of biosensors [1,2,3,4,5]. Functionalized nanostructured materials are used as carriers for nanobiocatalyst, which is a nanocarrier-enzyme composite material. These include organic-inorganic hybrids, mesoporous/nanoporous carriers, nanoparticles, magnetic nanoparticles, carbon nanotubes, and nanoflowers (HNFs) [6]. Nanoparticles attract much attention among many nanomaterials because they have many unique properties. The large surface area due to its morphology is just one of its unique features, which makes its activity and stability excellent [7,8,9].
Metal-containing nanoparticles are used in many scientific applications due to their small size and metal content [10,11,12]. Hybrid nanoflowers (hNFs), among the nanoparticles developed in recent years, have a flower-like structure and have many superior properties compared to other nanoparticles due to this morphology [13].
Hybrid nanoflowers were discovered accidentally by Zare and his group in 2012 through the self-assembling and co-precipitation of enzymes [14]. HNFs have been widely investigated since 2012 due to their good stability, enhanced activity, and easy and environmentally friendly preparation [15]. Hybrid nanoflowers are flower-like three-dimensional (3D) nanostructures of organic and inorganic components [16]. Hybrid nanoflowers are flower-like, providing a large surface-to-volume ratio and excellent catalytic activity. Recent studies have also preferred it due to its high stability [17].
Various metal ions such as Ca2+, Cu2+, Co2+, Fe2+, Mn2+, and Zn2+ are used as inorganic ingredients to form hybrid nanoflowers. Various biomolecules such as amino acids, DNA, enzymes, and organic molecules are preferred as organic ingredients [18,19,20].
There are many studies on hybrid nanoflowers today. For example, Güven et al. (2022) synthesized organic@inorganic hybrid copper nanoflowers (Cu NFs) using cherry stem for the first time. Cherry stem extract was obtained using ethanol and water solvents. Various techniques have been used to characterize Cu NFs by synthesis under different pH conditions. Cu NFs were revealed to have catalytic, antioxidant, and antimicrobial activities [21]. Koca and colleagues (2024) synthesized hNF using algae extract and Cu metal ion. They evaluated the effect of ambient pH and algal extract concentration on their morphology. Dye degradation and antioxidant activities of synthesized hNFs were determined under optimum conditions [22].
In the current study, for the synthesis of hNFs, 1,2-phenylenediamine, and N1-phenylbenzene-1,2-diamine were used as the organic part, while copper metal ion was used as the inorganic part. This was the first time hNFs were synthesized using the mentioned organic molecules. The characterization of 1,2-phenylenediamine@CuhNFs and N1-phenylbenzene-1,2-diamine@CuhNFs was conducted with FESEM, EDX, XRD, FT-IR, and elemental mapping. In addition, the cytotoxic effects of 1,2-phenylenediamine@CuhNFs and N1-phenylbenzene-1,2-diamine@CuhNFs were performed on A549 and MCF7 cells. The current study aims to create novel nanoflowers N1-phenylbenzene-1,2-diamine and 1,2-phenylenediamine compounds and analyze their activity on anticancer. Support was obtained from molecular docking to confirm the potential of these compounds for both breast and lung cancer. Molecular docking studies can provide information about binding interactions between compounds with identified targets, which may explain their inhibitory activity. A better result can be obtained by examining the binding patterns in the active binding site of the compounds through molecular docking.
2 Materials and methods
2.1 Chemicals and Reagents
CuSO4·5H2O KH2PO4, CaCl2·2H2O, MgCl2.6H2O, NaCl, KCl, and Na2HPO4 were provided from Sigma-Aldrich. N1-phenylbenzene-1,2-diamine and 1,2-phenylenediamine were purchased from BLD pharm and Merck, respectively.
2.2 Synthesis of N1-phenylbenzene-1,2-diamine@CuhNFs and 1,2-phenylenediamine@CuhNFs
N1-phenylbenzene-1,2-diamine@CuhNFs and 1,2-phenylenediamine@CuhNFs were synthesized using a method reported by Somturk et al. [23,24,25]. Three hundred thirty-three microliters of copper sulfate solution and also 1 mL of organic molecules (0.02 mg/mL) are added into 50 mL of PBS buffer (pH:7.4). After the mixture is gently vortexed, it is incubated at + 4 oC for three days. The mixture is centrifuged at 10,000 rpm for 30 min, and the pellet part is left to dry.
2.3 Characterization of N1-phenylbenzene-1,2-diamine@CuhNFs and 1,2-phenylenediamine@CuhNFs
Using the SEM device, it was realized that the synthesized N1-phenylbenzene-1,2-diamine@CuhNFs and 1,2-phenylenediamine@CuhNFs had a flower structure. Additionally, the presence of metal ions in the hNFs was elucidated using EDX. While XRD was used to elucidate the crystal structures of hNFs, the FTIR spectrum was used to determine the presence of phosphorus-dependent oxygen peaks. The average size of hNFs was determined using the Image ProPlas 6.0 program.
2.4 Cytotoxicity Assessment
MTT ([3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide]) test was used to reveal the cytotoxic effects of hNFs. For this, A549 and MCF7 cell lines were used. After the cells were increased, hybrid nanoflowers synthesized in the 15,625–1000 µg/mL concentration range were added to the wells and left for one night. Staining was performed, and cell viability was determined by measuring absorbance in a microplate reader. Statistical analysis was performed by one-way ANOVA.
2.5 Computational Studies
The results of the computational studies were obtained with the Schrödinger 2021-2 program [26]. The interactions of compounds N1-phenylbenzene-1,2-diamine and 1,2-phenylenediamine, which was determined to be effective as an anticancer according to in vitro results, on both EGFR and Human Estrogen Receptor were investigated via molecular docking. ADME properties were calculated to evaluate the pharmacokinetic behavior of the compounds.
2.5.1 Molecular Docking Studies
Molecular docking analyses were performed to investigate the active interaction modes of the compounds 1,2-phenylenediamine and N1-phenylbenzene-1,2-diamine interacting with the crystal structures (PDB IDs:1M17 [27], 3ERT [28]) obtained from the protein database. Targets important in cancer signaling pathways were identified for use in this study. Since the human lung cancer cell line and the human breast cancer cell line studied in vitro, targets are selected accordingly. In this study, EGFR was preferred for lung cancer, and estrogen receptor for breast cancer. In addition, in silico interactions of the designed compounds are essential because these targets prevent cancer cells from spreading throughout the body and metastasis. Ligand preparation, receptor preparation, and ligand-receptor in silico interaction were calculated following the procedure applied in previous studies [29,30,31].
Possible conformations of ligands are calculated with the Schrödinger 2021-2 LigPrep “Ligand preparation wizard” [32]. The crystal structure was prepared with the “Protein Preparation Wizard” of Schrödinger 2021-2 Protein Preparation [33] software. OPLS_2005 force field was used to perform optimization and minimization operations to prepare targets. While the prepared ligands and targets were labeled, their free binding energies were calculated with the MM/GBSA VSGB solvent model at the Schrödinger Prime wizard [34]. This analysis was applied to determine the free binding energies of the interaction results of 1,2-phenylenediamine and N1-phenylbenzene-1,2-diamine compounds with the target.
2.5.2 ADME Prediction
Absorption, distribution, metabolism, and excretion (ADME) properties were estimated with Schrödinger’s QikProp [35] wizard. The compounds were analyzed according to ADME predictions using the QikProp [35] tool of the Schrödinger program. QikProp values of these compounds, such as molecular weight, dipole moment, SASA, FISA, PISA, QPpolarize, QPlogPo/w, QPlogHERG, QPPCaco, QPlogBB, #metab, QPlogKhsa, human absorption percentage, PSA, violations of Lipinski rules (rule of five, rule of three) were calculated.
3 Results and Discussion
3.1 Characterization of hNFs
3.1.1 SEM Analysis
The morphologies of the synthesized hNFs were determined by FESEM analysis, and the images were determined to have flower-like structures. The average dimensions for CuhNFs were ∼ 8.13 μm and ∼ 9.46 μm, respectively (Fig. 1). As seen in the figure, it has been proven that the structures have a flower-like structure. This flower-shaped morphology causes the surface area to expand, which increases its stability and activity.
3.1.2 EDX Analysis
Elemental analysis of N1-phenylbenzene-1,2-diamine@CuhNFs and 1,2-phenylenediamine@CuhNFs were carried out with EDX. The EDX spectrum of CuhNFs is seen in Fig. 2. The presence of Cu metal ion in the copper phosphate nanocrystal can be seen in the figure.
3.1.3 Elemental Mapping
It is seen in Figs. 3 and 4 that the metal ions contained in hNFs are distributed homogeneously.
3.1.4 XRD Analysis
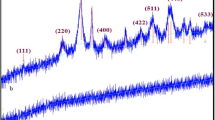
The crystal structure of the 1,2-phenylenediamine@CuhNFs and N1-phenylbenzene-1,2-diamine@CuhNFs were carried out with XRD (Fig. 5). The XRD peaks were assigned as for 1,2-phenylenediamine@CuhNFs and N1-phenylbenzene-1,2-diamine@CuhNFs, XRD: 8.93°, 12.82°, 18.47°, 20.54°, 22.78°, 26.66°, 29.56°, 33.80°, 35.31°, 36.04°, 37.28°, 38.44°, 41.98°, 47.05°, 53.55°, 55.29°, 57.17°, 61.79°, 65.19°, 68.43°, 72.68° in Fig. 5 compared with JCPDS (00 − 022 − 0548).
3.1.5 FT-IR Analysis
The chemical structure of 1,2-phenylenediamine@CuhNFs and N1-phenylbenzene-1,2-diamine@CuhNFs were illuminated using the FT-IR spectrum (Fig. 6). The spectrum data of 1,2-phenylenediamine@CuhNFs was seen as follows; For hNFs, FT-IR (cm− 1): 3305 (N–H and O–H stretching), 2975 (Ar–H and C–H stretching), 2912 (C–H, stretching), 1645, 1535, 1420, 1250, 1045 (P = O), 1055, 994 (P–O), 970, 683 (O = P = O), 634, 566 (O = P = O) and N1-phenylbenzene-1,2-diamine@CuhNFs were seen as follows; For hNFs, FT-IR (cm− 1): 3351 (N–H and O–H stretching), 2976 (Ar–H and C–H stretching), 2914 (C–H, stretching), 1631, 1542, 1430, 1350, 1059 (P = O), 1071, 994 (P–O), 952, 664(O = P = O), 637, 563 (O = P = O).
3.1.6 Cytotoxicity Assessment
The effect of ligands, 1,2-phenylenediamine@CuhNFs and N1-phenylbenzene-1,2-diamine@CuhNFs on the viability of A549 (lung) and MCF7 (breast) cancer cell lines was shown in Fig. 7. Different concentrations (15,625–1000 µg/mL) were applied to cells.
1,2-Phenylenediamine and 1,2-phenylenediamine@CuhNFs were tested in the A549 cell line. It was observed that the anticancer activity was better when the organic molecule was converted into a hybrid nanoflower. While the hybrid nanoflower caused 70% cell death, the organic molecule caused 40%. When the N1-phenylbenzene-1,2-diamine molecule A549 cell line was examined, it was determined that the hybrid nanoflower killed the cells by 90%, while the organic molecule killed by 60%. Although these two organic molecules were effective in the A549 cell line, it was observed that their effectiveness increased even more when they were converted into a hybrid nanoflower form. When the 1,2-phenylenediamine molecule was examined in the MCF7 cell line, it was observed that its anticancer activity was better when the organic molecule was converted into a hybrid nanoflower. While the hybrid nanoflower caused 70% cell death, it was observed that organic molecules did not affect the cell line. When the N1-phenylbenzene-1,2-diamine molecule was examined in the MCF7 cell line, it was determined that the hybrid nanoflower killed the cells by 80%, while the organic molecule killed by 40%. Although these two organic molecules were effective in the MCF7 cell line, it was observed that their effectiveness increased even more when they were converted into a hybrid nanoflower form. There are many studies in the literature about hybrid nanoflowers. However, there are only a few studies on anticancer. One is a study conducted by Somturk Yılmaz and his colleagues in 2024. This study conducted a cytotoxic study on the A49 cell line with different metal ions using Tribulus Terrestris L. plant extract. They observed that its anticancer activity increased significantly when the extract was converted into a hybrid nanoflower form [36]. Recent studies have shown that, in addition to proteins and enzymes (containing amino groups), it has been proven in many studies that hybrid nanoflowers can be synthesized with plant extracts and organic molecules (containing carboxyl and hydroxyl groups).
3.2 Computational Studies
3.2.1 Molecular Docking Studies
Molecular docking studies have been performed to investigate the ligand-protein binding pattern and orientations within binding sites on anticancer targets. However, both ligands docked to the active binding region of the crystal structures of both the breast cancer target (PDB ID:3ERT [28]) and the lung cancer target (PDB ID:1M17 [27]) obtained from the protein database. Table 1 presents the computational results of the interaction of these crystal structures with both ligands.
In silico approaches to support in vitro studies with computational methods, firstly, the results of the N1-phenylbenzene-1,2-diamine, and 1,2-phenylenediamine compounds interacting with the crystal structure of the human estrogen receptor (PDB ID: 3ERT) were analyzed. In Table 1, the docking score values of N1-phenylbenzene-1,2-diamine, and 1,2-phenylenediamine compounds were calculated as -7.304 kcal/mol and − 4.157 kcal/mol, respectively. The result of these calculations was also visualized to analyze the binding mode. Table 1 also presents the result of the binding energy values of the second target, the Epidermal Growth Factor Receptor. Docking score values between EGFR and both ligands were calculated with the Schrödinger 2021-2 Glide program. The binding interaction values between EGFR and compounds N1-phenylbenzene-1,2-diamine and 1,2-phenylenediamine were calculated as -4.220 kcal/mol, -3.596 kcal/mol, respectively.
Support was taken from Prime MM/GBSA analysis to investigate the relative binding affinity of the two ligands with both targets. Within the binding site of the 3ERT crystal structure, two compounds (N1-phenylbenzene-1,2-diamine, 1,2-phenylenediamine) showed good binding free energy values (ΔGBind) of − 55.03 kcal/mol, − 32.69 kcal/mol, respectively. The second target, calculating the binding free energy of the ligands with the 1M17 crystal structure, resulted in ΔGBind values of -50.06 kcal/mol, -29.20 kcal/mol for N1-phenylbenzene-1,2-diamine, 1,2-phenylenediamine, respectively. All MM-GBSA calculations are summarized in Table 1. Both ligands’ calculated binding affinity (ΔGBind) is different because they bind to their active sites with different amino acids while binding to both targets. As a result, docking score values and free binding energy values are consistent with biological values.
In Figs. 2D, 3D and 8 interaction diagrams of the interactions of N1-phenylbenzene-1,2-diamine, 1,2-phenylenediamine compounds with 1M17, the EGFR crystal structure, are presented. The 2D interaction diagram of the N1-phenylbenzene-1,2-diamine compound located in the active pocket region of the 1M17 crystal structure and the hydrogen bond interaction with Asp831, one of the essential amino acids is presented in Fig. 8(A). The 3D representation of the N1-phenylbenzene-1,2-diamine-1M17 complex is presented in Fig. 8(B). In the representation in Fig. 8(B), it is observed that critical amino acids surround the N1-phenylbenzene-1,2-diamine compound.
As a result of its in silico interaction with the EGFR target, the second ligand, 1,2-phenylenediamine, interacted with Asp831, a critical amino acid in this target, through hydrogen bonding, as mentioned before. The visual of this interaction is presented in Fig. 8(C). Figure 8(D) presents a 3D visual of the 1,2-phenylenediamine compound.
(A) 2D and (B) 3D docking pose displaying interaction of compound N1-phenylbenzene-1,2-diamine in the binding site of Epidermal Growth Factor Receptor (PDB ID: 1M17). (C) 2D and (D) 3D docking poses displaying the interaction of compound 1,2-phenylenediamine in the binding site of the Epidermal Growth Factor Receptor (PDB ID: 1M17)
The crystal structure of the target used as an estrogen receptor in breast cancer is presented due to the interaction of 3ERT and the ligands specified in Table 1. 2D and 3D interaction diagrams of ligands interacting with the 3ERT target are presented in Fig. 9. It is presented in Fig. 9(A) that the expected hydrogen bond interaction of the N1-phenylbenzene-1,2-diamine compound with the amino acid residues Glu353 and Luu346, which are essential in this target, occurs. It is understood from the 3D interaction in Fig. 9(B) that the N1-phenylbenzene-1,2-diamine compound can interact with the 3ERT crystal structure by settling in the active pocket site.
It is understood from the images in Fig. 9(C) and Fig. 9(D) that the 1,2-phenylenediamine compound binds to the estrogen receptor in the correct area, and the expected hydrogen bonds occur. The hydrogen bond interaction of the compound 1,2-phenylenediamine with the amino acid residues Leu387 and Glu353 caused the ligand to settle into the active binding region correctly.
When the molecular docking results were evaluated quantitatively and qualitatively, both ligands were analyzed in silico and in vitro. The binding parameter values presented in Table 1 are pretty striking. Moreover, Figs. 8 and 9 present this to us qualitatively.
3.2.2 ADME Profiling Studies
ADME calculation of compounds that may be drug candidates using in silico approaches is a frequently used prediction tool. ADME parameter values were calculated using Schrödinger 2021-2 Qikprop wizard in silico approaches. Determining the physicochemical and pharmacokinetic properties of ligands is an essential tool. Therefore, to check the drug-likeness of the N1-phenylbenzene-1,2-diamine and 1,2-phenylenediamine compounds, physicochemical properties were performed using Schrödinger 2021-2 Qikprop (Table 2).
The ADME profile and Lipinski Rule values of both ligands are presented in Table 2. It was determined that the ADME parameters of the compounds were within the acceptable range for human use. The values in Table 2 present the positive pharmacokinetic profile of the compounds N1-phenylbenzene-1,2-diamine and 1,2-phenylenediamine. The results are presented in Table 2. Lipinski’s rule of five (Ro5) parameters are calculated to estimate the probability of drug similarity when deciding whether the synthesized compounds will be drug candidates. According to Lipinski’s rule of three and five, all values are acceptable. Both ADME pharmacokinetics and Lipinski’s Rule values indicate that this compound is expected to have drug-like properties and has the potential to be an orally active drug. Elangovan and colleagues (2023) synthesized and characterized the Schiff base from 3,4,5-trimethoxybenzaldehyde and sulfadiazine. A Swiss-ADME was used for a pharmacological study to determine whether the TMBDA molecule has pharmacological properties. The synthesized compound was found to show good antimicrobial activity compared to standard [37]. Elangovan et al. (2023) reported a Schiff base synthesized from vanillin and o-phenylenediamine. Electronic spectra, HOMO-LUMO and MESP, were performed with the gas and various solvent phases [38]. Elangovan and colleagues (2023) synthesized the (E)-3-(((9H-purin-6-yl)imino)methyl)phenol (A3H) molecule and characterized its structure with spectroscopic techniques. The molecule’ structure was optimized using the B3LYP/cc-pVDZ basis set. The electronic properties of molecule was analyzed and they found that water has the lowest energy gap value compared to other solvents. Molecular docking techniques determined the binding interaction between the targeted proteins and the ligand [39].
4 Conclusion
To summarize, hNF synthesis was conducted using 1,2-phenylenediamine and N1-phenylbenzene-1,2-diamine molecules as the organic fraction and Cu (II) metal ion as the inorganic fractions. A549 and MCF7 cell lines were used to determine the cytotoxic effects of the synthesized hybrid nanoflowers. The compound N1-phenylbenzene-1,2-diamine plays a vital role in the localization of both targets in the binding pocket site according to findings from molecular docking studies. It causes a significant interaction due to forming a critical hydrogen bond with ASp831 at the EGFR target. In the second target, the estrogen receptor, both ligands settled in the active binding site with the support of hydrogen bonding to interact with Glu353 and Leu346. Due to both ADME profile, docking score, and binding mode values, ligands are suitable for further development as compounds that could be anticancer drug candidates.
Data Availability
No datasets were generated or analysed during the current study.
References
A.O. Dada, F.A. Adekola, F.E. Dada, A.T. Adelani-Akande, M.O. Bello, C.R. Okonkwo, A.A. Inyinbor, A.P. Oluyori, A. Olayanju, K.O. Ajanaku, Adetunji C.O. Silver nanoparticle synthesis by Acalypha wilkesiana extract: phytochemical screening, characterization, influence of operational parameters, and preliminary antibacterial testing. Heliyon 5(10), e02517 (2019)
B. Mahdavi, S. Saneei, M. Qorbani, M. Zhaleh, A. Zangeneh, M.M. Zangeneh, E. Pirabbasi, N. Abbasi, H. Ghaneialvar, Ziziphora clinopodioides Lam leaves aqueous extract mediated synthesis of zinc nanoparticles and their antibacterial, antifungal, cytotoxicity, antioxidant, and cutaneous wound healing properties under in vitro and in vivo conditions. Appl. Organomet. Chem. 33, e5164 (2019)
S.A. Khan, F. Noreen, S. Kanwal, A. Iqbal, G. Hussain, Green synthesis of ZnO and Cu-doped ZnO nanoparticles from leaf extracts of Abutilon indicum, Clerodendrum Infortunatum, Clerodendrum inerme and investigation of their biological and photocatalytic activities. Mater. Sci. Eng. : C 82, 46–59 (2018)
E. Mostafa, M.A.A. Fayed, R.A. Radwan, R.O. Bakr, Centaurea Pumilio L. extract and nanoparticles: a candidate for healthy skin. Colloids Surf. B 182, 110350 (2019)
N.A. Sumbal, S. Naz, J.S. Ali, A. Mannan, M. Zia, Synthesis, characterization and biological activities of monometallic and bimetallic nanoparticles using Mirabilis jalapa leaf extract. Biotechnol. Rep. 22, e00338 (2019)
L. Hernandez-Morales, H. Espinoza-Gomez, L.Z. Flores-Lopez, E.L. Sotelo-Barrera, A. Nunez-Rivera, R.D. Cadena-Nava, G. Alonso-Nunez, K.A. Espinoza, Study of the green synthesis of silver nanoparticles using a natural extract of dark or white Salvia hispanica L. seeds and their antibacterial application. Appl. Surf. Sci. 489, 952–961 (2019)
F. Shao, A.J. Yang, D.M. Yu, J. Wang, X. Gong, H.X. Tian, Bio-synthesis of Barleria Gibsoni leaf extract mediated zinc oxide nanoparticles and their formulation gel for wound therapy in nursing care of infants and children. J. Photochem. Photobiol B 189, 267–273 (2018)
E. Pinon-Segundo, N. Mendoza-Munoz, D. Quintanar-Guerrero, Nanoparticles as dental drug-delivery systems. Nanobiomater Clin. Dent. 475–495 (2013)
S. Mukherjee, D. Chowdhury, R. Kotcherlakota, S.B.V. Patra, M.P. Bhadra, B. Sreedhar, C.R. Patra, Potential theranostics application of bio-synthesized silver nanoparticles (4-in-1 system). Theranostics. 4(3), 316–335 (2014)
S. Sonia, J.K.H. Linda, K. Ruckmani, M. Sivakumar, Antimicrobial and antioxidant potentials of biosynthesized colloidal zinc oxide nanoparticles for a fortified cold cream formulation: a potent nanocosmeceutical application. Mater. Sci. Eng. C 79, 581–589 (2017)
R. Mukhopadhyay, J. Kazi, M.C. Debnath, Synthesis and characterization of copper nanoparticles stabilized with Quisqualis indica extract: evaluation of its cytotoxicity and apoptosis in B16F10 melanoma cells. Biomed. Pharmacother. 97, 1373–1385 (2018)
K. Mio, R. Stern, Inhibitors of Hyaluronidases. Matrix Biol. 21, 31–37 (2002)
O.T. Gül, I. Ocsoy, Preparation of magnetic horseradish peroxidase-laccase nanoflower for rapid and efficient dye degradation with dual mechanism and cyclic use. Mater. Lett. 303, 130501 (2021)
J. Ge, J. Lei, R.N. Zare, Protein-inorganic hybrid nanoflowers. Nat. Nanotechnol. 7, 428–432 (2012)
S. Dayan, C. Altinkaynak, Ş.D. Doğan, N. Kayaci, N. Ozdemir, Hybrid nanoflowers bearing tetraphenylporphyrin assembled on copper (II) or cobalt (II) inorganic material: a green efficient catalyst for hydrogenation of nitrobenzenes in water. Appl. Organomet. Chem. 34, 5381 (2020)
J. Gao, H. Liu, C. Tong, L. Pang, Y. Feng, M. Zuo et al., Hemoglobin-Mn3(PO4)2 hybrid nanoflower with opulent electroactive centers for high-performance hydrogen peroxide electrochemical biosensor. Sens. Actuators B Chem. 307, 127628 (2020)
S.K.S. Patel, S. Otari, J. Li, D.R. Kim, S.C. Kim, B.-K. Cho et al., Synthesis of cross-linked protein-metal hybrid nanoflowers and its application in repeated batch decolorization of synthetic dyes. J. Hazard. Mater. 347, 442–450 (2018)
Y.-K. Luo, F. Song, X.-L. Wang, Y.-Z. Wang, Pure copper phosphate nanostructures with controlled growth: a versatile support for enzyme immobilization. Cryst. Eng. Comm. 19, 2996–3002 (2017)
D. Kong, R. Jin, X. Zhao, H. Li, X. Yan, F. Liu et al., Protein–inorganic hybrid nanoflower-rooted agarose hydrogel platform for point-of-care detection of acetylcholine. ACS Appl. Mater. Interfaces. 11, 11857–11864 (2019)
G. He, W. Hu, C.M. Li, Spontaneous interfacial reaction between metallic copper and PBS to form cupric phosphate nanoflower and its enzyme hybrid with enhanced activity. Colloids Surf. B: Biointerfaces. 135, 613–618 (2015)
O.C. Güven, M. Kar, F.D. Koca, Synthesis of Cherry Stalk Extract based Organic@Inorganic Hybrid Nanofowers as a Novel Fenton reagent: evaluation of their antioxidant, Catalytic, and Antimicrobial activities. J. Inorg. Organomet. Polym. Mater. 32, 1026–1032 (2022)
F.D. Koca, H.M. Muhy, M.G. Halici, Catalytic and antioxidant activity of Desmarestia Menziesii Algae Extract Based Organic@İnorganic Hybrid Nanofowers. J. Inorg. Organomet. Polym. Mater. 34, 1282–1292 (2024)
B. Somturk, I. Yilmaz, C. Altinkaynak, A. Karatepe, N. Özdemir, I. Ocsoy, Synthesis of urease hybrid nanoflowers and their enhanced catalytic properties. Enzyme Microb. Technol. 86, 134–142 (2016)
B. Somturk, S. Dayan, N. Ozdemir, Kalaycıoğlu Özpozan N. Catalytic performance improvement with metal ion changes for efficient, stable, and reusable superoxide dismutase–metalphosphates hybrid nanoflowers. Chem. Pap. 76, 4245–4260 (2022)
B. Somturk, M. Hançer, İ. Öcsoy, N. Ozdemir, Synthesis of copper ion incorporated horseradish peroxidase based hybrid nanoflowers for enhanced catalytic activity and stability. Dalton Trans. 44(31), 13845–13852 (2015)
Schrödinger, Release 2021-2: Glide, S (LLC, New York, NY, 2021)
J. Stamos, M.X. Sliwkowski, C. Eigenbrot, Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J. Biol. Chem. 277(48), 46265–46272 (2002)
A.K. Shiau, D. Barstad, P.M. Loria, L. Cheng, P.J. Kushner, D.A. Agard, G.L. Greene, The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 95(7), 927–937 (1998)
İ.B. Merde, G.T. Önel, B. Türkmenoğlu, Ş. Gürsoy, E. Dilek, Pyridazinones containing the (4-methoxyphenyl) piperazine moiety as AChE/BChE inhibitors: design, synthesis, in silico and biological evaluation. Med. Chem. Res. 31(11), 2021–2031 (2022)
İ. BozbeyMerde, G.T. Önel, B. Türkmenoğlu, Ş. Gürsoy, E. Dilek, (p-Chlorophenyl)‐3 (2H) pyridazinone derivatives: synthesis, in Silico, and AChE/BChE inhibitory activity. ChemistrySelect. 7(38), e202202446 (2022)
B. Türkmenoğlu, Investigation of novel compounds via in silico approaches of EGFR inhibitors as anticancer agents. J. Indian Chem. Soc. 99(8), 100601 (2022)
Schrödinger, Release 2021-2: LigPrep, S (LLC, New York, NY, 2021)
Schrödinger, S. Epik, LLC, New York, NY, 2021
Schrödinger, Release 2021-2: Prime, S (LLC, New York, NY, 2021)
Schrödinger, Release 2021-2: Qikprop, L., New York, NY, 2021
B. Somturk Yilmaz, H. Bekci, A. Altiparmak, S. Uysal, İ. Şenkardeş, G. Zengin, Determination of anticancer activity and biosynthesis of Cu, Zn, and Co hybrid nanoflowers with Tribulus terrestris L. extract. Process. Biochem. 138, 14–22 (2024)
N. Elangovan, S.Y. Alomar, S. Sowrirajan, B. Rajeswari, A. Nawaz, A.N. Kalanthoden, Photoluminescence property and solvation studies on (E)-N-(pyrimidin-2yl)-4-((3,4,5-trimethoxy benzylidede) amino) benzene sulfonamide; synthesis, structural, topological analysis, antimicrobial activity, and molecular docking studies. Inorg. Chem. Commun. 155, 111019 (2023)
N. Elangovan, S. Ganesan, T. Vignesh, R. Alomar, S.Y. Sowrirajan, S. Chandrasekar, S. Kalanthoden, A.N. Nawaz, Influence of the solvents through molecular structure, wavefunction studies, biological activity prediction and molecular docking studies of 4,4-((1,2-phenylenebis (azaneylylidene)) bis (methaneylylidene)) bis(2-methoxyphenol). J. Mol. Liq. 391, 123334 (2023)
N. Elangovan, S. Sowrirajan, N. Arumugam, A.I. Almansour, S.M. Mahalingam, S. Kanchana, Synthesis, solvent role (water and DMSO), antimicrobial activity, reactivity analysis, inter and intramolecular charge transfer, topology, and molecular docking studies on adenine derivative. J. Mol. Liq. 391, 123250 (2023)
Acknowledgements
The authors would like to thank Erzincan Binali Yıldırım University, Basic Sciences Application and Research Center (EBYU-EUTAM) for the Schrödinger Maestro 2021-2 program. The authors thank Erciyes University, Drug Application and Research Center (ERFARMA) for laboratory and device support. SA would like to thank the Suleyman Demirel University Research Fund (TSG-2021-8458) for financial support.
Funding
Open access funding provided by the Scientific and Technological Research Council of Türkiye (TÜBİTAK). Süleyman Demirel University Research Fund provided partial financial support for purchasing the starting material with the project code TSG-2021-8458.
Open access funding provided by the Scientific and Technological Research Council of Türkiye (TÜBİTAK).
Author information
Authors and Affiliations
Contributions
B.S.Y., B.T., and S.A. performed all experiments. B.S.Y., B.T., and S.A. conceived the original idea and designed the project. S.A. wrote the manuscript.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Somturk-Yilmaz, B., Turkmenoglu, B. & Akkoc, S. Synthesis, Characterization, Cytotoxic Activity Studies of N1-phenylbenzene-1,2-diamine @CuhNFs and 1,2-phenylenediamine@CuhNFs, and Molecular Docking Calculations of Their Ligands. J Inorg Organomet Polym (2024). https://doi.org/10.1007/s10904-024-03178-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10904-024-03178-1