Abstract
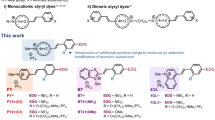
The properties of six commonly used, commercially available, fluorescent dyes were compared in staining right-handed B-DNA and left-handed Z-DNA. All showed different degree of fluorescence turn-on in the presence of B-DNA, but very little in the presence of Z-DNA. The optimal range of dye-DNA ratios of DNA was determined. While these dyes do not provide a turn-on type probe for Z-DNA, staining between B- and Z-DNA using dyes such as SYBR Green I was shown to be useful in tracking the kinetics of conformational changes between these two forms of DNA. Finally, SYBR Green I showed unique circular dichroism patterns in 4 M NaCl that change in the presence of double stranded DNA, both in the visible and UV range.











Similar content being viewed by others
Data Availability
Not applicable.
References
ThermoFisher (2010) The Molecular Probes Handbook, the 11th Ed
Huang Q, Fu WL (2005) Comparative analysis of the DNA staining efficiencies of different fluorescent dyes in preparative agarose gel electrophoresis. Clin Chem Lab Med 43(8):841–842
Haines AM, Tobe SS, Kobus HJ, Linacre A (2015) Properties of nucleic acid staining dyes used in gel electrophoresis. Electrophoresis 36(6):941–944
Yan X, Grace WK, Yoshida TM, Habbersett RC, Velappan N, Jett JH, Keller RA, Marrone BL (1999) Characteristics of different nucleic acid staining dyes for DNA fragment sizing by flow cytometry. Anal Chem 71(24):5470–5480
Specht EA, Braselmann E, Palmer AE (2017) A critical and comparative review of fluorescent tools for live-cell imaging. Annu Rev Physiol 79:93–117
Gudnason H, Dufva M, Bang DD, Wolff A (2007) Comparison of multiple DNA dyes for real-time PCR: effects of dye concentration and sequence composition on DNA amplification and melting temperature. Nucleic Acids Res 35(19):e127
Jovin T, Soumpasis DM, McIntosh LP (1987) The transition between B-DNA and Z-DNA. Annu Rev Phys Chem 38:521–560
Rich A, Zhang S (2003) Z-DNA: the long road to biological function. Nat Rev Genet 4(7):566–572
Yan H, Powers R, Gibbons A, Joshi D (2017) Z-DNA: Chemistry and Biological Relevance. In Elsevier Reference Module in Chemistry, Molecular Sciences and Chemical Engineering; Reedijk, J., Ed.; Elsevier: Waltham, MA
Fuertes MA, Cepeda V, Alonso C, Peŕez JM (2006) Molecular mechanisms for the B-Z transition in the example of poly[d(G-C)•(G-C)] polymers. A critical review. Chem Rev 106(6):2045–2064
Solodinin A, Gautrais A, Ollivier S, Yan H (2019) Incorporation of 5-fluoro-2’-deoxycytidine into oligonucleotides for the study of DNA structures by 19F NMR spectroscopy. ACS Omega 4:19716–19722
Biver T, García B, Leal JM, Secco F, Turriani E (2010) Left-handed DNA: intercalation of the cyanine thiazole orange and structural changes. A kinetic and thermodynamic approach. Phys Chem Chem Phys 12:13309–13317
Dumat B, Larsen AF, Wilhelmsson LM (2016) Studying Z-DNA and B- to Z-DNA transitions using a cytosine analogue FRET-pair. Nucleic Acids Res 44(11)
Aeschbacher M, Reinhardt CA, Zbinden GA (1986) Rapid cell membrane permeability test using fluorescent dyes and flow cytometry. Cell Biol Toxicol 2(2):247–255
Cosa G, Focsaneanu KS, McLean JRN, McNamee JP, Scaiano JC (2004) Photophysical properties of fluorescent DNA-dyes bound to single- and double-stranded DNA in aqueous buffered solution. Photochem Photobiol 73(6):585–599
Chiaraviglio L, Kirby JE (2014) Evaluation of impermeant, DNA-binding dye fluorescence as a real-time readout of eukaryotic cell toxicity in a high throughput screening format. Assay Drug Dev Technol 12(4):219–228
Mao F, Leung WY, Xin X (2007) Characterization of EvaGreen and the implication of its physicochemical properties for qPCR applications. BMC Biotechnol 7(76):1–16
Briggs C, Jones M (2005) SYBR Green I-induced fluorescence in cultured immune cells: A comparison with Acridine Orange. Acta Histochem 107(4):301–312
Guzaev M, Li X, Park C, Leung WY, Roberts L (2017) Comparison of nucleic acid gel stains. Cell permeability, safety, and sensitivity of ethidium bromide alternatives. Biotium
Le Pecq JB (1971) Use of ethidium bromide for separation and determination of nucleic acids of various conformational forms and the measurement of their associated enzymes. In: Glick D (ed) Methods of Biochemical Analysis, vol 20. JohnWiley and Sons, New York, pp 41–86
Saeidnia S, Abdollahi M (2013) Are other fluorescent tags used instead of ethidium bromide safer? DARU J Pharm Sci 21(1):71
Fuller W, Waring MJ (1964) A molecular model for the interaction of ethidium bromide with deoxyribonucleic acid. Ber Bunsenges Phys Chem 68(8–9):805–808
Nicoletti I, Migliorati G, Pagliacci MC, Grignani F, Riccardi C (1991) A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J Immunol Methods 139(2):271–279
Boulos L, Prévost M, Barbeau B, Coallier J, Desjardins R (1999) LIVE/DEAD® BacLightTM: application of a new rapid staining method for direct enumeration of viable and total bacteria in drinking water. J Microbiol Methods 37(1):77–86
Tsai CC, Jain SC, Sobell HM (1977) Visualization of drug-nucleic acid interactions at atomic resolution: I Structure of an ethidium/dinucleoside monophosphate crystalline complex, ethidium: 5-iodouridylyl (3′–5′) adenosine. J Mol Biol 114(3):301–315
Jain SC, Tsai CC, Sobell HM (1977) Visualization of drug-nucleic acid interactions at atomic resolution: II Structure of an ethidium/dinucleoside monophosphate crystalline complex, ethidium: 5-iodocytidylyl (3′–5′) guanosine. J Mol Biol 114(3):317–331
Jain SC, Sobell HM (1984) Visualization of drug-nucleic acid interactions at atomic resolution: VIII Structures of two ethidium/dinucleoside monophosphate crystalline complexes containing ethidium: cytidylyl(3′-5′) guanosine. J Biomol Struct Dyn 1(5):1179–1194
Kapuscinski J (1995) DAPI: a DNA-specific fluorescent probe. Biotech Histochem 70(5):220–233
Shoute LCT, Loppnow GR (2018) Characterization of the binding interactions between EvaGreen dye and dsDNA. Phys Chem Chem Phys 20:4772–4780
Beaudet MP, Cox GW, Yue S (2005) Molecular Probes, Inc., USA. WO/2005/033342
Evenson WE, Boden LM, Muzikar KA, O’Leary DJ (2012) 1H and 13C NMR assignments for the cyanine dyes SYBR Safe and Thiazole Orange. J Org Chem 77(23):10967–10971
Pei R, Rothman J, Xie Y, Stojanovic MN (2009) Light-up properties of complexes between thiazole orange-small molecule conjugates and aptamers. Nucleic Acids Res 37(8):e59
Jin X, Yue S, Wells KS, Singer VL (1994) SYBR™ Green I: a new fluorescent dye optimized for detection of picogram amounts of DNA in gels. Biophys J 66:A159
Zipper H, Brunner H, Bernhagen J, Vitzthum F (2004) Investigations on DNA intercalation and surface binding by SYBR Green I, its structure determination and methodological implications. Nucleic Acids Res 32(12):e103
Dragan AI, Pavlovic R, McGivney JB, Casas-Finet JR, Bishop ES, Strouse RJ, Schenerman MA, Geddes CD (2012) SYBR Green I: fluorescence properties and interaction with DNA. J Fluoresc 22(4):1189–1199
Alaranta JM, Truong K-N, Matus MF, Malola SA, Rissanen KT, Shroff SS, Marjomäki VS, Häkkinen HJ, Lahtinen TM (2023) Optimizing the SYBR green related cyanine dye structure to aim for brighter nucleic acid visualization. Dyes Pigments 208:110844
Hur JH, Lee AR, Yoo W, Lee JH, Kim KK (2019) Identification of a new Z-DNA inducer using SYBR green 1 as a DNA conformation sensor. FEBS Lett 593(18):2628–2636
Hannah C, Armitage BA (2004) DNA-templated assembly of helical cyanine dye aggregates: A supramolecular chain polymerization. Acc Chem Res 37(11):845–853
Funding
This work was funded by the Natural Sciences and Engineering Research Council of Canada, and National Research Council of Canada.
Author information
Authors and Affiliations
Contributions
HB: writing – original draft, methodology, investigation, formal analysis. AM: review & editing, methodology. HY: Conceptualization, supervision, formal analysis, writing, review & editing.
Corresponding author
Ethics declarations
Ethical Approval
Not applicable.
Conflict of Interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bennett, HA., McAdorey, A. & Yan, H. Staining Properties of Selected Commercial Fluorescent Dyes Toward B- and Z-DNA. J Fluoresc 34, 1193–1205 (2024). https://doi.org/10.1007/s10895-023-03343-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10895-023-03343-8