Abstract
Using computer simulation we investigated whether machine learning (ML) analysis of selected ICU monitoring data can quantify pulmonary gas exchange in multi-compartment format. A 21 compartment ventilation/perfusion (V/Q) model of pulmonary blood flow processed 34,551 combinations of cardiac output, hemoglobin concentration, standard P50, base excess, VO2 and VCO2 plus three model-defining parameters: shunt, log SD and mean V/Q. From these inputs the model produced paired arterial blood gases, first with the inspired O2 fraction (FiO2) adjusted to arterial saturation (SaO2) = 0.90, and second with FiO2 increased by 0.1. ‘Stacked regressor’ ML ensembles were trained/validated on 90% of this dataset. The remainder with shunt, log SD, and mean ‘held back’ formed the test-set. ‘Two-Point’ ML estimates of shunt, log SD and mean utilized data from both FiO2 settings. ‘Single-Point’ estimates used only data from SaO2 = 0.90. From 3454 test gas exchange scenarios, two-point shunt, log SD and mean estimates produced linear regression models versus true values with slopes ~ 1.00, intercepts ~ 0.00 and R2 ~ 1.00. Kernel density and Bland–Altman plots confirmed close agreement. Single-point estimates were less accurate: R2 = 0.77–0.89, slope = 0.991–0.993, intercept = 0.009–0.334. ML applications using blood gas, indirect calorimetry, and cardiac output data can quantify pulmonary gas exchange in terms describing a 20 compartment V/Q model of pulmonary blood flow. High fidelity reports require data from two FiO2 settings.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
More than 50 years ago John West published his landmark model of pulmonary gas exchange [1], building on the work of predecessors [2]. The model is characterised by volumes of inspired gas (V) and mixed venous blood (Q) equilibrating in 10 to 100 virtual lung compartments governed by log normal distributions of alveolar ventilation and pulmonary capillary blood flow across compartmental V/Q ratios [1, 3, 4].
The multiple inert gas technique (MIGET), an investigative tool based on West’s model [5, 6], has provided mechanistic detail on impaired gas exchange. MIGET evaluations are technically challenging procedures in which six inert gases spanning a range of solubilities are infused in saline until equilibration. Plots of pulmonary retention and excretion versus gas solubility are constructed from gas chromatographic measurements and ‘transformed’ respectively into distributions of blood flow and ventilation against a logarithmic scale of V/Q ratios spread across 50 compartments [5, 7].
MIGET has identified shunt (V/Q = 0) as the dominant cause of hypoxaemia in the acute respiratory distress syndrome (ARDS) and lobar pneumonia, whereas in chronic obstructive pulmonary disease (COPD) and in some patients with COVID-19 pneumonia hypoxaemia is primarily from mixed venous equilibration in low V/Q compartments [8,9,10]. Bimodal distributions have been observed in patients with COPD, asthma [3] and ARDS [11].
Despite its ‘gold standard’ status, the complexity of MIGET has obliged clinicians to track pulmonary gas exchange via alternative indices, usually those categorized as ‘tension’ or ‘content’-based [12]. Venous admixture (VA) is the classic content-based index [13], while tension-based indices include the A–a gradient, used in APACHE risk algorithms [14], and the ratio between the arterial oxygen tension and the inspired oxygen fraction (PaO2/FiO2 ratio or PF ratio), important in ARDS diagnosis and stratification [15].
These indices show significant signal variability [16], but their greatest drawback is the limited information provided on the underlying pulmonary pathophysiology. The VA approach of Riley and Cournand [13, 17] is more informative on this aspect, but hampered by inherent over-simplification. This is because VA (V/Q = 0) is one of just two perfused compartments (V/Q = 0 and 1). All oxygen transfer deficits are corralled within VA, in other words as true shunt, leaving no ability to tease out contributions from low V/Q compartments. For clinicians this can be a crucial distinction, for example in managing COVID-19 pneumonia (see “Discussion” section) [10]. Similarly, the effects of high V/Q are incorporated in a single dead space estimate (V/Q = ∞). As a final drawback, accurate VA calculations require mixed venous blood for analysis [12].
In part to address these shortcomings, scaled back variations on the MIGET framework have been proposed [18,19,20,21]. Prominent among these is the automatic lung parameter estimator (ALPE) [18], described as a ‘simple bedside alternative to MIGET’. ALPE has been shown to match complex MIGET calculations in experimental lung injury [22, 23], and is now finding application in clinical research [24] and as the key component of a commercial package (www.mermaidcare.com) designed for monitoring and decision support.
Like MIGET, shunt is given conventionally in ALPE assessments as percentage of cardiac output. However, unlike MIGET, ALPE models ‘low’ and ‘high’ V/Q mismatch as partial pressure differentials (to be distinguished from diffusion limitation) across imposed ‘partitions’ between blood and alveolar gas. Specifically, ‘low’ V/Q mismatch is represented by the fall in PO2 from alveolar gas to pulmonary end-capillary blood, and ‘high’ V/Q mismatch as the rise in PCO2 across the same interface.
We suggest that machine learning (ML) could add value in this ‘scaled back MIGET’ space [25, 26]. With data inputs close to those used by ALPE it should be possible for trained ML applications to generate detailed pulmonary assessments. These could take the form of a shunt estimate plus separate parameters defining log normal distributions of blood flow across compartmental V/Q ratios. Critical care physicians would then be provided with prompt actionable diagnostic information presented in a familiar format. Added bonuses could include shorter measurement intervals with a reduced requirement for FiO2 ‘switching’ (at present ALPE requires up to four FiO2 ‘switches’).
To investigate this possibility, we tested the following hypotheses in silico:
-
(1)
Trained ML applications using data normally sourced from blood gas analysis, indirect calorimetry, and cardiac output measurements can quantify pulmonary gas exchange in terms describing a multi-compartment V/Q model of pulmonary blood flow.
-
(2)
Consistent ML reports require measurement data at no more than two FiO2 settings.
2 Materials and methods
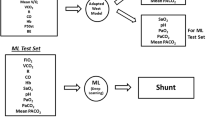
To test the above hypotheses, we exposed selected ML applications to simulated clinical monitoring data routinely available from blood gas analysis, indirect calorimetry, and cardiac output measurements. Scenarios were constructed with these data to represent a diverse mix of O2 consumption (VO2) and delivery, CO2 production (VCO2) and transport, hemoglobin-oxygen affinity, and respiratory and metabolic acid–base status. Paired blood gases were generated in each simulation by a 21-compartment model of pulmonary blood flow governed by three input values: shunt percentage, log standard deviation (log SD) and distributional mean (Fig. 1, for more model detail and core equations, see Supplementary Material).
Graphical illustration of modelled blood flow through 20 gas exchanging compartments plus a single shunt compartment (V/Q = 0). Shunt is set at 10% of total pulmonary blood flow. Note the log normal distribution of the non-shunt pulmonary blood flow according to compartment V/Q ratios. In this example log SD = 2.0 and flow distributional V/Q mean = 0.35
To make the evaluation, ML applications trained on this material were challenged with simulated monitoring data from ‘unseen’ test scenarios, the goal in each case being to back-generate the three governing model parameters of pulmonary blood flow distribution (shunt, log SD and mean). These estimates were then compared with ‘true’ model input values for the same scenarios.
Steps in this process were as follows:
-
(1)
Arterial blood gases were produced by the lung model at two structured settings of inspired oxygen fraction (FiO2) (see below) in response to unique input combinations of the three parameters defining model pulmonary blood flow distribution (shunt, log SD and mean, Table 1) plus one value from each of six monitoring categories (Table 2) available from blood gas analysis, indirect calorimetry, and cardiac output measurements.
Table 1 Model defining parameters Table 2 Monitoring inputs with ranges -
(2)
Using a Python program, 34,551 unique input combinations were built around a core set of 7500.
-
(3)
Model calculations were run from VBA sub-routines (Excel, Microsoft, Redmond, WA) until stable outputs were achieved for pH, PCO2, PO2 and Hb saturation in arterial and mixed venous blood and in the pulmonary end-capillary blood of each of the 20 non-shunt compartments.
-
(4)
For each input combination, the FiO2 generating an arterial oxygen saturation (SaO2) of 0.90 was determined by iteration, ensuring that in each case 0.21 ≤ FiO2 ≤ 0.90.
-
(5)
On attainment of SaO2 = 0.90, values were logged for FiO2, arterial pH, arterial PO2 (PaO2), arterial PCO2 (PaCO2), calculated PF ratio and calculated venous admixture (VA).
-
(6)
For the second calculation the FiO2 was increased by 0.1 and the model run again.
-
(7)
Values for SaO2 and calculations of VA, and PF ratios were logged at this higher FiO2.
-
(8)
With data from SaO2 = 0.90 as baseline, changes at the higher FiO2 in SaO2 (Dsat), VA (DVA) and PF ratios (DPF) were calculated and logged.
-
(9)
This sequence performed 34,551 times generated the final dataset.
2.1 ML analysis of completed dataset
-
(1)
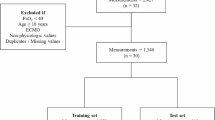
After pre-processing to reduce redundancies, data rows were formatted as in Table 3 and subjected to randomization.
-
(2)
The randomized dataset was partitioned into sequential split fractions (70%:20%:10%) for ML training, validation and testing respectively.
-
(3)
The test fraction was subjected to trained ML analysis with columns containing the model-defining values of pulmonary blood flow (shunt, log SD and mean) ‘held back’ to allow blinded estimates.
-
(4)
Two categories of ML estimates were performed:
-
(a)
‘Single-Point’ estimates were derived by ML analysis of 10 variables confined to model input and output logs for SaO2 = 0.90. Input variables were ‘CO2load’, ‘O2pull’, standard P50 (P50st) [27], base excess, BE [28], and blood haemoglobin concentration (Hb). Output variables were FiO2, arterial pH, PaCO2, PaO2, and VA (Table 3).
-
(a)
-
(b)
‘Two-Point’ estimates were derived after inclusion of three additional variables consisting of DVA, Dsat and DPF (Table 3), all obtained from model output logs following the 0.10 FiO2 increment.
2.2 ML methodology
We used open-source ML algorithms implementing linear regression techniques [Supplementary Material Table 1(s)]. It became evident during the validation process that multiple simultaneous models in a ‘stacked’ or ‘ensemble’ configuration outperformed any single model. The stacking process used simple linear regression at the output layer to combine the contributions from individual models.
Model stacks were tested using ‘StackingRegressor’ from the ‘sklearn’ Python library (https://scikit-learn.org/stable/). Models were trained using correlation (‘R’ and ‘R2), mean absolute error (‘MAE’) and by comparing the slope and distance from zero intersection of the line of best fit.
See ‘Supplementary Material’ for more detail of ML methodologies employed.
2.3 Statistical analysis
Prior to analysis, the comparison data were checked for completeness, accuracy, and consistency.
Two-way (univariate) comparisons were made using standard linear regression. Post-estimation diagnostics were run on all models. Due to the large size of the dataset, these included checking model residuals for normality, using both the Kolmogorov–Smirnov test and a normal probability plot and heteroskedasticity, using the Breusch–Pagan and Cook–Weisberg tests. For each predictor, the regression slope (β) and its p-value were tabulated along with the equation intercept and the overall R2 value.
Kernel density plots and graphical Bland and Altman analyses [29] were constructed to enable visual comparisons of single-point and two-point results for each variable (shunt, log SD, and mean estimates) versus the true values.
STATATM (v17.0) was used for all analyses with the level of significance set throughout at α < 0.05.
3 Results
From the final dataset of 34,551 data rows, 31,097 rows were allocated for ML training and validation and the remaining 3454 rows for testing.
From the 3454-row test-set, kernel density and Bland and Altman plots of single-point and two-point estimates by ML versus true values of shunt, log SD and mean are set out in Figs. 2, 3, 4, 5, 6 and 7. All distributions are non-normal. Corresponding regression data are reported in Table 4, and Bland and Altman data in Table 5.
Shunt (single-point). Two subplots are illustrated. The Bland–Altman (BA) plot illustrates the 3454 points. For clarity, each point is horizontally jittered by ± 1% of the value of the independent variable. Horizontal plot lines indicate the median and 95% confidence interval for the difference (enumerated in Table 5). The kernel density estimate (KDE) plot illustrates the distribution of observations for the independent variable. The solid line is the true value of the variable with the dashed line indicating the modeled variable. Each subplot shares the same X-axis scale. Both X-axis units and the Y-axis units in the BA plot are defined by the independent variable. The Y-axis in the KDE plot is dimensionless
Shunt (two-point). Description as for Fig. 2
Mean (single-point). Description as for Fig. 2
Mean (two-point). Description as for Fig. 2
Log SD (single-point). Description as for Fig. 2
Log SD (two-point). Description as for Fig. 2
3.1 Two-point estimates
Two-point estimates of shunt, log SD and mean produced regression models with almost identical results (Table 4), with β ~ 1.00, intercept ~ 0.00 and R2 ~ 1.00 for each of the test-set variables. The kernel density and Bland and Altman plots confirmed close agreement with true values (Figs. 3, 5, 7; Table 5).
3.2 Single-point estimates
From Figs. 2, 4 and 6 and Tables 4 and 5, single-point estimates showed close concordance but less consistent reflections of true values. Ranges from the regression models of the three estimate categories versus true values were R2 = 0.77–0.89, β = 0.991–0.993, and intercepts = 0.009–0.334 (Table 4).
4 Discussion
Using computer simulation, we found that blinded ML analysis of monitoring data replicating diverse gas exchange scenarios, including blood gases generated by a 21-compartment V/Q model of pulmonary blood flow, could back-generate the model’s governing parameters. This was achieved with ‘stacked regressor’ ML ensembles trained and tested on blood gas, indirect calorimetry, and cardiac output data over a broad spectrum of gas exchange equilibria. In each simulation ML accurately delineated pulmonary blood flow as shunt percentage plus the key descriptors (log SD and mean) of log normal flow distributions to gas exchanging compartments according to their V/Q ratios. This is essentially pulmonary blood flow in MIGET format.
Measurements adopted for the simulation are available from current ICU monitoring devices [30]. Point of care blood gas analysis has been routine in ICU practice for decades. Indirect calorimetry is now recommended as a nutritional guide for critically ill mechanically ventilated patients [31,32,33]. Low invasive cardiac output monitoring, although not without problems [34,35,36], is mainstream in contemporary ICUs. The application of artificial intelligence in critical illness monitoring and decision support is itself no longer a novel concept [26].
The dataset to train, validate and test the ML applications was derived from systematically varied input combinations of the three model defining parameters (shunt, log SD, and mean, Table 1), linked to four direct measurements (cardiac output, VO2, VCO2, and Hb; Table 2) and two calculated parameters (BE, P50st; Table 2). To complete each scenario the model generated paired sets of arterial blood gases in response to these inputs at two structured FiO2 settings. The final dataset represented approximately 35,000 unique scenarios covering a diverse mix of O2 delivery and consumption, CO2 production and transport, hemoglobin-oxygen affinity, and respiratory and metabolic acid–base status.
ML was then able to back-generate the model-defining parameters of 3454 test scenarios in blinded fashion using only the blood gas measurements along with inherent derived values (BE, P50st, VA, PF ratios) plus cardiac output, VO2, VCO2, and the baseline FiO2. ML estimates from single-point data (recorded at baseline SaO2 = 0.90) showed sufficient concordance with true values to reflect trends in all three key model parameters. However, a second equilibration introduced a dynamic component, captured by ML via changes in VA (DVA), PF ratios (DPF) and saturation (Dsat). This two-point approach enabled high fidelity identification of all three key model descriptors (Figs. 3, 5, 7; Tables 4, 5).
The simulation was designed to emulate a practical two-step procedure in which arterial blood gas analysis with oximetry is performed with the FiO2 adjusted for SaO2 = 0.90 (using SpO2 as initial guide). This is followed by a second set of blood gases after increasing the FiO2 by 0.10. During this process, once only measurements of cardiac output, VO2 and VCO2 are also recorded. ML then quantifies the defining parameters of the diagnostic model(s) of choice from relationships embedded in the data.
It should be possible to train ML applications in other diagnostic models such as the ALPE system, which like the approach considered here devolves to three key parameters [18, 37], in that case shunt and partial pressure gradients across modelled blood/gas ‘partitions’ representing ‘high V/Q’ and ‘low V/Q’ mismatch. It is also conceivable that larger training datasets with wider input ranges could enable accurate single-point ML reports from data ‘snapshots’ collected at any working FiO2. One further possibility for future investigation is that training sets formatted to target specific model variants, for example bimodal flow distributions [38], could extend ML reporting to these complexities.
Informative ‘on the spot’ gas exchange evaluations can facilitate management decisions, as mentioned in the Introduction. A contemporary example might be a ventilated patient with pneumonia and hypoxemia with a PF ratio < 100. To decide on a safe course of action clinicians should be able to distinguish between two extremes of lung pathophysiology. At one extreme the disturbed oxygenation represents a large right to left shunt in the context of low pulmonary gas volumes, typical of recruitable ARDS. At the other pulmonary gas volumes are normal and shunt is minimal, the hypoxemia arising instead from widespread low V/Q ratios due to maldistributed lung perfusion, a situation more characteristic of COVID-19 with multiple pulmonary vascular thrombi. In the latter circumstance, recruitment maneuvers and major manipulations of positive end expiratory pressure (PEEP) would be contraindicated [10, 38]. Varying combinations of the two extremes complete the spectrum of possibilities.
Based on our simulation, ML evaluations could make these distinctions rapidly without a need for specialized imaging. Equivalent diagnostic assessment by the current ALPE system would take 10 to 15 min, involve up to four FiO2 ‘switches’, and report VQ mismatch as partial pressure gradients [24, 37].
4.1 Some caveats
The model of pulmonary blood flow used to generate the blood gases follows the basic West model format. Several modifications and simplifications were employed. These are detailed in the Supplementary Material.
The simulation assumes error-free measurements, whereas some degree of error is intrinsic to measurements of cardiac output [36], indirect calorimetry [39], and the measured and derived elements of blood gas analysis [12]. Indirect calorimetry has increased error potential at FiO2 ≥ 0.7 or PEEP > 12, both encountered in severe respiratory failure [39]. Other risk factors include circuit leaks, bronchopleural fistulae, and possibly extracorporeal circulations.
We have not attempted a sensitivity analysis. However, it is noteworthy that ALPE, an advanced system now in service, is subject to similar error susceptibilities. ALPE evaluations require a single arterial blood gas analysis and one cardiac output measurement or estimate, along with measurements at three to five different FiO2 settings of VO2, VCO2, arterial oxygen saturation by pulse oximetry (SpO2), and end-tidal O2 and CO2 fractions [24]. Despite measurement intervals of 10–15 min with up to four FiO2 ‘switches’, any signal distortion from absorption atelectasis [40] and altered hypoxic pulmonary vasoconstriction [41] is regarded as minor [37].
Further, the MIGET gold-standard itself relies on a series of measurements and techniques all prone to error, including but not limited to cardiac output and minute ventilation measurements, collection of mixed expired gas without condensation-induced loss of dissolved gases, and gas chromatographic concentration measurements of six inert gases in both mixed expired gas and the gas phases above blood samples [4].
The low baseline arterial saturation (SaO2 = 0.90) was selected to allow a subsequent 0.10 FiO2 step-up within the bounds of FiO2 ≤ 1.00. Although SaO2 = 0.90 is at the hypoxemia threshold [12], it is considered adequate for tissue oxygenation in the absence of anemia and low cardiac output, albeit with limited supportive evidence [42]. Of historical interest, older versions of the automated ALPE system could manipulate baseline SaO2 to values as low as 0.85, if necessary using FiO2 < 0.21 [18].
Dataset shunt, log SD and mean values retained uneven distributions across their respective ranges, as illustrated by the test-set kernel density plots (Figs. 2, 3, 4, 5, 6, 7). Greater training set uniformity may have produced more consistent single-point estimations. Barriers to uniformity included the automatic rejection of input combinations in which SaO2 ≠ 0.90 when FiO2 ≥ 0.21 ≤ 0.90.
5 Conclusions
We conclude based on computer simulations of diverse gas exchange scenarios that trained ML applications using data sourced from blood gas analysis, indirect calorimetry, and cardiac output measurements can quantify pulmonary gas exchange in terms used to describe multi-compartment V/Q models of pulmonary blood flow. High fidelity ML reports require measurement data at no more than two FiO2 settings, subject to measurement accuracy.
References
West JB. Ventilation–perfusion inequality and overall gas exchange in computer models of the lung. Respir Physiol. 1969;7(1):88–110.
Farhi LE, Rahn H. A theoretical analysis of the alveolar-arterial O2 difference with special reference to the distribution effect. J Appl Physiol. 1955;7(6):699–703.
West JB. State of the art: ventilation–perfusion relationships. Am Rev Respir Dis. 1977;116(5):919–43.
West JB, Wagner PD. Pulmonary gas exchange. In: West JB, editor. Bioengineering aspects of the lung. New York: Marcel Dekker; 1977. p. 361–457.
Wagner PD. The multiple inert gas elimination technique (MIGET). Intensive Care Med. 2008;34(6):994–1001.
Wagner PD, Laravuso RB, Uhl RR, West JB. Continuous distributions of ventilation–perfusion ratios in normal subjects breathing air and 100 per cent O2. J Clin Investig. 1974;54(1):54–68.
Yu G, Yang K, Baker AB, Young I. The effect of bi-level positive airway pressure mechanical ventilation on gas exchange during general anaesthesia. Br J Anaesth. 2006;96(4):522–32.
D’Alonzo GE, Dantzker DR. Respiratory failure, mechanisms of abnormal gas exchange, and oxygen delivery. Med Clin N Am. 1983;67(3):557–71.
Rodriguez-Roisin R, Roca J. Mechanisms of hypoxemia. Intensive Care Med. 2005;31(8):1017–9.
Gattinoni L, Gattarello S, Steinberg I, Busana M, Palermo P, Lazzari S, et al. COVID-19 pneumonia: pathophysiology and management. Eur Respir Rev. 2021. https://doi.org/10.1183/16000617.0138-2021.
Dantzker DR, Brook CJ, Dehart P, Lynch JP, Weg JG. Ventilation–perfusion distributions in the adult respiratory distress syndrome. Am Rev Respir Dis. 1979;120(5):1039–52.
Morgan TJ, Venkatesh B. Monitoring oxygenation. In: Bersten AD, Handy JM, editors. Oh’s intensive care manual. 8th ed. Philadelphia: Butterworth-Heinemann Elsevier; 2018. p. 160–70.
Riley RL, Cournand A. Analysis of factors affecting partial pressures of oxygen and carbon dioxide in gas and blood of lungs; theory. J Appl Physiol. 1951;4(2):77–101.
Zimmerman JE, Kramer AA, McNair DS, Malila FM. Acute Physiology and Chronic Health Evaluation (APACHE) IV: hospital mortality assessment for today’s critically ill patients. Crit Care Med. 2006;34(5):1297–310.
Force ADT, Ranieri VM, Rubenfeld GD, Thompson BT, Ferguson ND, Caldwell E, et al. Acute respiratory distress syndrome: the Berlin Definition. JAMA. 2012;307(23):2526–33.
Kathirgamanathan A, McCahon RA, Hardman JG. Indices of pulmonary oxygenation in pathological lung states: an investigation using high-fidelity, computational modelling. Br J Anaesth. 2009;103(2):291–7.
Riley RL, Cournand A, Donald KW. Analysis of factors affecting partial pressures of oxygen and carbon dioxide in gas and blood of lungs; methods. J Appl Physiol. 1951;4(2):102–20.
Rees SE, Kjaergaard S, Perthorgaard P, Malczynski J, Toft E, Andreassen S. The automatic lung parameter estimator (ALPE) system: non-invasive estimation of pulmonary gas exchange parameters in 10–15 minutes. J Clin Monit Comput. 2002;17(1):43–52.
Loeppky JA, Caprihan A, Altobelli SA, Icenogle MV, Scotto P, Vidal Melo MF. Validation of a two-compartment model of ventilation/perfusion distribution. Respir Physiol Neurobiol. 2006;151(1):74–92.
Vidal Melo MF, Loeppky JA, Caprihan A, Luft UC. Alveolar ventilation to perfusion heterogeneity and diffusion impairment in a mathematical model of gas exchange. Comput Biomed Res. 1993;26(2):103–20.
Lockwood GG, Fung NL, Jones JG. Evaluation of a computer program for non-invasive determination of pulmonary shunt and ventilation–perfusion mismatch. J Clin Monit Comput. 2014;28(6):581–90.
Rees SE, Kjaergaard S, Andreassen S, Hedenstierna G. Reproduction of MIGET retention and excretion data using a simple mathematical model of gas exchange in lung damage caused by oleic acid infusion. J Appl Physiol (1985). 2006;101(3):826–32.
Rees SE, Kjaergaard S, Andreassen S, Hedenstierna G. Reproduction of inert gas and oxygenation data: a comparison of the MIGET and a simple model of pulmonary gas exchange. Intensive Care Med. 2010;36(12):2117–24.
Karbing DS, Panigada M, Bottino N, Spinelli E, Protti A, Rees SE, et al. Changes in shunt, ventilation/perfusion mismatch, and lung aeration with PEEP in patients with ARDS: a prospective single-arm interventional study. Crit Care. 2020;24(1):111.
Sidey-Gibbons JAM, Sidey-Gibbons CJ. Machine learning in medicine: a practical introduction. BMC Med Res Methodol. 2019;19(1):64.
Burki TK. Artificial intelligence hold promise in the ICU. Lancet Respir Med. 2021. https://doi.org/10.1016/S2213-2600(21)00317-9.
Siggaard-Andersen O, Siggaard-Andersen M, Fogh-Andersen N. The TANH-equation modified for the hemoglobin, oxygen, and carbon monoxide equilibrium. Scand J Clin Lab Investig Suppl. 1993;214:113–9.
Siggaard-Andersen O. The Van Slyke equation. Scand J Clin Lab Investig Suppl. 1977;37(146):15–20.
Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1(8476):307–10.
Morgan TJ, Anstey CM. Expanding the boundaries of point of care testing. J Clin Monit Comput. 2020;34(3):397–9.
Singer P, Pichard C, Rattanachaiwong S. Evaluating the TARGET and EAT-ICU trials: how important are accurate caloric goals? Point-counterpoint: the pro position. Curr Opin Clin Nutr Metab Care. 2020;23(2):91–5.
De Waele E, Honore PM, Malbrain M. Does the use of indirect calorimetry change outcome in the ICU? Yes it does. Curr Opin Clin Nutr Metab Care. 2018;21(2):126–9.
Singer P, Blaser AR, Berger MM, Alhazzani W, Calder PC, Casaer MP, et al. ESPEN guideline on clinical nutrition in the intensive care unit. Clin Nutr. 2019;38(1):48–79.
Saugel B, Cecconi M, Wagner JY, Reuter DA. Noninvasive continuous cardiac output monitoring in perioperative and intensive care medicine. Br J Anaesth. 2015;114(4):562–75.
Monnet X, Teboul JL. Minimally invasive monitoring. Crit Care Clin. 2015;31(1):25–42.
Teboul JL, Saugel B, Cecconi M, De Backer D, Hofer CK, Monnet X, et al. Less invasive hemodynamic monitoring in critically ill patients. Intensive Care Med. 2016;42(9):1350–9.
Karbing DS, Kjaergaard S, Andreassen S, Espersen K, Rees SE. Minimal model quantification of pulmonary gas exchange in intensive care patients. Med Eng Phys. 2011;33(2):240–8.
Busana M, Giosa L, Cressoni M, Gasperetti A, Di Girolamo L, Martinelli A, et al. The impact of ventilation–perfusion inequality in COVID-19: a computational model. J Appl Physiol (1985). 2021;130(3):865–76.
Lev S, Cohen J, Singer P. Indirect calorimetry measurements in the ventilated critically ill patient: facts and controversies—the heat is on. Crit Care Clin. 2010;26(4):e1-9.
Dantzker DR, Wagner PD, West JB. Proceedings: instability of poorly ventilated lung units during oxygen breathing. J Physiol. 1974;242(2):72P.
Grant BJ, Davies EE, Jones HA, Hughes JM. Local regulation of pulmonary blood flow and ventilation–perfusion ratios in the coatimundi. J Appl Physiol. 1976;40(2):216–28.
Gilbert-Kawai ET, Mitchell K, Martin D, Carlisle J, Grocott MP. Permissive hypoxaemia versus normoxaemia for mechanically ventilated critically ill patients. Cochrane Database Syst Rev. 2014. https://doi.org/10.1002/14651858.cd009931.pub2.
Zou GY. Confidence interval estimation for the Bland–Altman limits of agreement with multiple observations per individual. Stat Methods Med Res. 2013;22(6):630–42.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. Project supported by Departmental Funds.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Morgan, T.J., Langley, A.N., Barrett, R.D.C. et al. Pulmonary gas exchange evaluated by machine learning: a computer simulation. J Clin Monit Comput 37, 201–210 (2023). https://doi.org/10.1007/s10877-022-00879-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10877-022-00879-1