Abstract
The title compound, C22H28F3N3OS2+ × 2(C6H2N3O7 −) × (CH3)2SO—the picrate salt of the potent antipsychotic drug, fluphenazine—crystallizes in the triclinic P-1 space group with unit cell parameters a = 10.6333(12) Å, b = 11.9696(12) Å, c = 17.7036(15) Å, α = 103.265(9)°, β = 98.414(9)°, γ = 102.702(10)°. The ionic fragments: the fluphenazinium dictation, and two picrate anions, are joined by means of strong N–H···O and weak C–H···O hydrogen bonds into the chains of alternating cations and anions, expanding along [010] direction. Within the chain, the picrates interact by means of short π···π interactions: the mean distance between the planes is 3.366 Å; additional interaction of the same type between one of the picrates and the phenyl ring of the phenothiazine ring system additionally strengthen the pattern. The phenothiazine ring exists in a typical, “butterfly-like” conformation, with two terminal rings planar and the central ring in a slightly flattened boat form. This conformation results in the dihedral angle between the terminal rings of 41.76(5)°. The aliphatic chain which substitutes phenothiazine at N-position is not in an extended conformation, the torsion angles along this chain are 75.95(14)° and −163.96(10)°. The structure contains also the solvent—dimethylsulphoxide molecules, which are connected with the cation–anion structure by means of strong O–H···O hydrogen bonds.
Graphical Abstract
The ionic fragments of the title salt are joined by means of strong N–H···O and weak C–H···O hydrogen bonds into the chains of alternating cations and anions, expanding along [010] direction

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Experimental
Preparation
Fluphenazine dihydrochloride (5.6 g, 0.01 mol) was dissolved in 25 mL of methanol and picric acid (2.4 g, 0.01 mol) was dissolved in 25 mL of methanol. Both the solutions were mixed and the mixture was stirred in a beaker at 333 K for 1 h and was kept aside for 3 days at room temperature. The formed salt was filtered and dried in vaccum desiccator over phosphorous pentoxide. It was observed that, regardless of the proportions in which the donor and acceptor were mixed, only the 1:2 complex was formed. X-ray quality crystals were obtained from a solution of dimethylsulphoxide after slow evaporation (m.p.: 501–503 K).
Crystallography
Yellow transparent prism-like crystal (0.2, 0.2, 0.08 mm) was used for data collection. Diffraction data were collected at 120(1) K by the ω-scan technique up to 2θ = 56.5°, on an Xcalibur Eos diffractometer [1] with graphite-monochromatized MoKα radiation (λ = 0.71073 Å). The temperature was controlled by an Oxford Instruments Cryosystems cooling device. The data were corrected for Lorentz-polarization and absorption effects [1]. Accurate unit-cell parameters were determined by a least-squares fit of 14624 reflections of highest intensity, chosen from the whole experiment. The structure was solved with SIR92 [2] and refined with the full-matrix least-squares procedure on F2 by SHELXL97 [3] Scattering factors incorporated in SHELXL97 were used. The function Σw(∣Fo∣2 − ∣Fc∣2)2 was minimized, with w−1 = [σ2(Fo)2 + (0.0513P)2 + 0.13P] (where P = [Max (F 2o , 0) + 2F 2c ]/3). All non-hydrogen atoms were refined anisotropically, hydrogen atoms were found in the difference Fourier map, and refined with the isotropic displacement parameters set at 1.2 (1.5 for methyl groups) times the Ueq value for appropriate non-hydrogen atom. Relevant crystal data are listed in Table 1, together with refinement details.
CCDC-775478 contains supplementary crystallographic data for this paper. These data can be obtained free of charge via www.ccdc.cam.ac.uk/data_request/cif, by e-mailing data_request@ccdc.cam.ac.uk, or by contacting The Cambridge Crystallographic Data Centre, 12 Union Road, Cambridge CB2 1EZ, UK.
Results and Discussion
Fluphenazine (2-[4-[3-[2-(trifluoromethyl)-10H-phenothiazin-10-yl]propyl]-piperazin-1-yl]ethanol) belongs to the phenothiazine family of antipsychotic drugs used to treat schizophrenia and psychotic symptoms such as hallucinations, paranoia, and hostility. These drugs exert their activity by the blockade of dopaminergic receptors in the brain; they also inhibit alpha adrenoreceptors and muscarinic, cholinergic, histamine, serotonin, and opioid receptors in the central nervous system [e.g., 4, 5]. Fluphenazine is one of the piperazine class of phenothiazines and is extremely potent; more potent than haloperidol and around 50–70 times the potency of chlorpromazine [6]. It is marketed under the different brand names as decanoate (e.g., Modecate, Fludecate), enanthate (e.g., Flunanthate) or hydrochloride (e.g., Prolixin, Dapotum, Lyogen). The identification and differentiation of some phenothiazine picrates have been carried out [7] and a review on various aspects of phenothiazines has been published [8].
The structures of similar neutral molecules have been determined; for instance perphenazine (Cl instead of CF3, monoclinic Cc [9] and triclinic P1 [10] forms) and The crystal structures of some picrates of phenothiazines [11–15] have recently been reported. The present work reports the crystal structure of the salt formed by the interaction between 2-[4-[3-[2-(trifluoromethyl)-10H-phenothiazin-10-yl]propyl]-piperazin-1-yl] ethanol dihydrochloride and 2,4,6-trinitrophenol in methanol medium. The compound turned out to be a salt, fluphenazinium dipicrate and its crystal structure showed that it crystallizes as the dimethylsulphoxide solvate (Scheme 1).
The conformational preferences of phenothiazine ring system was described by Brock et al. [16]. Since then the number of structures deposited in the Cambridge Structural Database ([17]; Version of Nov. 2009) has grown substantially—the number of hits for phenothiazine is 151 (44 in [16])—but the observed preferences has not changed. Figure 1 presents the histogram of dihedral angles between the terminal ring planes for organic phenothiazine derivatives, without additional fused rings. The majority of the compounds have “butterfly-wings” conformation, with the dihedral angles between 25° and 45°.
The histogram showing the conformational preferences of the phenothiazine ring system in the compounds found in the CSD [14]. The plot shows the number of structures found versus the value of the dihedral angle between the planes of the terminal rings
Table 2 lists some selected geometrical parameters for 1. Figure 2 shows the anisotropic displacement representation of the dictation with labeling scheme. In 1, the phenothiazine ring system adopts the typical “butterfly-like” conformation (Fig. 2): the terminal phenyl rings are approximately planar (maximum deviations of 0.0257 (10) Å) while the central ring is a slightly flattened boat with the S and N atoms above the mean plane made by the remaining four carbon atoms. The asymmetry parameters [18], indicating the deviations from the ideal C 2v symmetry, have very low values: ΔC S1 s is 0.78° and ΔCs [2–7] is 2.44°. This conformation results in the dihedral angle between the terminal ring planes of 41.76 (5)°. This value is close to the mean value for the structures found in the CCDC (cf. Fig. 1). The C15 atom lies in equatorial position (C2–C7–N8–C15 torsion angle is 166.49 (12)°), and this equatorial plane is almost perpendicular to the plane defined by the rest of the bridging atoms (C15–C16–C17–N18 torsion angle is −163.96 (11)°), the dihedral angle is 82.06 (6)°. The piperazine ring is in the almost perfect chair conformation, maximum values of asymmetry parameters (in this case the ideal symmetry is D 3d ) are found for ΔC 20 s , of 0.94° and ΔC 2 [18, 19] of 1.2°). Both substituents of this ring are in equatorial positions (torsion angles C20–C19–N18–C17-177.21 (11)°, C19–C20–N21–C24-179.34 (11)°).
Anisotropic ellipsoid representation of the dictation together with atom labeling scheme [20]. The ellipsoids are drawn at 50% probability level, hydrogen atoms are depicted as spheres with arbitrary radii
The picrate anions have similar bond lengths and angles patterns, but their conformations are to some extent different. Figure 3 shows the anisotropic representation of both picrates with labeling schemes. The dihedral angles between almost planar ring and the planes of nitro groups can serve as descriptors of the conformation: in one of the anions (C47–O62) all three nitro groups are significantly twisted (dihedral angles are 36.05 (6)° and 23.24 (7)° for the groups ortho-with respect to the C=O(–) fragment and 20.78 (6)° for the para group). In the second anion the para group is closer to coplanarity, the dihedral angle is only 8.74 (2)°. Moreover, even though the two ortho groups are similarly twisted (26.45 (6)° and 24.43 (7)°), one of these nitro groups is twisted in opposite sense (cf. Fig. 3).
Anisotropic ellipsoid representations of the anions together with atom labeling scheme [20]. The ellipsoids are drawn at 50% probability level, hydrogen atoms are depicted as spheres with arbitrary radii
In the crystal structure there are hydrogen bonds connecting the structural elements of the structure (Table 3). Three strongest hydrogen bond donors, two N–H and one O–H groups from the cation are involved—as expected by Etter’s rules [19]—with the best acceptors: two C=O(–) oxygen atoms from picrate anions and carbonyl oxygen of the DMSO molecule (Fig. 4).
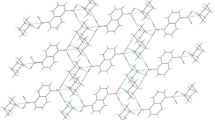
Fragment of the crystal packing as seen along b-direction, i.e., along the layer of the molecules [20]
A number of relatively short C–H···O contacts join the cations, anions and solvent molecules into complicated three-dimensional structure (cf. Table 3). The picrates from two neighbouring units connect two cations by alternating strong N–H···O and weak C–H···O hydrogen bonds into a hydrogen bonded dimer (Fig. 4). Within this dimmer, the relatively short π···π interactions between the picrate fragments and the picrate and one of the terminal rings of the phenothiazine system additionally stabilize this structure. The distance between the centers of the picrates is 3.471(1) Å, and between the picrate and phenyl—3.874 Å. Taking into account the offset, the interplanar distances are 3.366 and 3.525 Å, respectively. As might be seen from the Fig. 4, two different ‘sides’ of the cation take part in these interactions, so in the crystal structure these dimmers expand into the ribbons along b-direction (Fig. 5).
Fragment of the crystal packing as seen along b-direction, i.e., along the layer of the molecules [20]
References
Oxford Diffraction (2009) CrysAlis PRO (Version 1.171.33.36d). Oxford Diffraction Ltd
Altomare A, Cascarano G, Giacovazzo C, Gualardi A (1993) J Appl Cryst 26:343
Sheldrick GM (2008) Acta Cryst A64:112
Lieberman JA, Bymaster FP, Meltzer HY, Deutch AY, Duncan GE, Marx CE, Aprille JR, Dwyer DS, Li X-M, Mahadik SP, Duman RS, Porter JH, Modica-Napolitano JS, Newton SS, Csernansky JG (2008) Pharmacol Rev 60:358
Cieślik-Boczula K, Szwed J, Jaszczyszyn A, Gasiorowski K, Koll A (2009) J Phys Chem B113:15495
Martindale W (1977) In: Wade A (ed) The extra pharmacopoeia. The Pharmaceutical Press, London, pp 1567–1569
Yung DK, Pernarowski M (1963) J Pharm Sci 52:365
Kojlo A, Karpinska J, Kuzmicka L, Misiuk W, Puzanowska-Tarasiewicz H, Tarasiewicz M (2001) J Trace Microprobe Tech 19:45
McDowell JJH (1978) Acta Cryst B34:686
Lyle AJ, Wood C, Srikrishnan T (2008) J Chem Crystallogr 38:261
Yathirajan HS, Ashok MA, Narayana Achar B, Bolte M (2007) Acta Cryst E63:o1432
Yathirajan HS, Ashok MA, Narayana Achar B, Bolte M (2007) Acta Cryst E63:o1691
Yathirajan HS, Ashok MA, Narayana Achar B, Bolte M (2007) Acta Cryst E63:o1693
Yathirajan HS, Ashok MA, Narayana Achar B, Bolte M (2007) Acta Cryst E63:o1792
Yathirajan HS, Ashok MA, Narayana Achar B, Bolte M (2007) Acta Cryst E63:o1795
Brock CP, deLaLuz J, Golinski M, Lloyd MA, Vanaman TC, Watt DS (1996) Acta Cryst B52:713
Allen FH (2002) Acta Cryst B58:380
Duax WL, Norton DA (1975) Atlas of steroid structures. Plenum, New York, pp 16–22
Etter MC (1990) Acc Chem Res 23:120
Siemens (1989) Stereochemical workstation operation manual. Release 3.4. Siemens analytical X-ray instruments Inc. Madison, Wisconsin, USA
Acknowledgments
CSC thanks University of Mysore for research facilities.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Dutkiewicz, G., Siddaraju, B.P., Yathirajan, H.S. et al. The Crystal Structure of Fluphenazinium Dipicrate Dimethylsulphoxide Solvate. J Chem Crystallogr 40, 970–974 (2010). https://doi.org/10.1007/s10870-010-9773-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10870-010-9773-z