Abstract
3D structures and spectroscopic properties are reported of four compounds with the ligand bis(2-benzimidazolyl)ethane (hereafter abbreviated as dbz) attached to CuCl2, all having the general formula [Cu(dbz)Cl2](Hb) x . (in which Hb = EtOH or MeOH and x = 1/2 or 1). The X-ray crystal structure has been solved from these four slightly different compounds, namely: green α-[CuCl2dbz](C2H5OH)1/2 (1), the red compound β-[CuCl2dbz](C2H5OH)1/2 (2), which both have two slightly different units in the unit cell, the red α-[CuCl2dbz](CH3OH) (3) and a blue–green compound β-[CuCl2dbz](CH3OH) (4). The geometry around the Cu(II) anion is distorted tetrahedral for all four compounds, with chromophores consisting of two nitrogen atoms of the bidentate chelating dbz molecule and two chloride anions.
The unit cells of compound 1 and 2 consist of two chemically identical, but crystallographically different units, while compounds 3 and 4 each have only one independent CuCl2-(dbz) unit. The major differences are observed in the dihedral angles NCuN–ClCuCl, which vary from 29.3 to 77.1° for the four compounds. The differences are related to different packing effects, ring–ring stacking and H-bond interaction, due to the two different alcohols used. In fact these four compounds represent a new range of examples of distortion isomerism in pseudo-tetrahedrally coordinated species. Characterisation of the four compounds has been completed by IR, EPR and LF spectroscopy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The coordination chemistry of the heterocyclic azoles acting as ligands in copper(II) compounds in the context of modelling biological systems has gained much interest in the past decades [1–4]. In this respect bis(2-benzimidazoles) have received much attention for their wide-ranging antivirus activity [5, 6], their importance in selective ion-exchange resins [7, 8] and the possibility to form supramolecular aggregates with d10 metal ions [9].
So far, spectroscopic and magnetic properties, as well as X-ray structural analysis of a number of mono-, bis-, tetra- and polynuclear copper(II) coordination compounds with bis(2-benzimidazolyl)alkanes and some substituted bis(2-benzimidazolyl)alkanes have been reported by others and by us [10–20].
The copper(II) ion is known to yield in some cases different geometries with the same ligand. This has been coined as “plasticity effect” [21–23] and is believed to results from the Jahn–Teller distortion of the Cu(II) ion. Several ligands are known to yield different (coloured) Cu(II) isomers of the same compound, which have been classified as distortion isomers, a phenomenon known for many years [24–26].
In our earlier research with the ligand bis(2-benzimidazolyl)propane (abbreviated as tbz), two different Cu(II) compounds, of formula [Cu(tbz)Cl2], were obtained, i.e. a red isomer with a (distorted) tetrahedral geometry and a polymeric green isomer containing two different copper(II) sites, each distorted from square planar [14]. With a methyl substituent on different positions on the alkyl chain of tbz a number of copper(II) halide compounds were synthesized, however, no distortion isomers were reported in these cases [19].
In the present study a few other examples dealing with four CuCl2 distortion isomers are reported with the ligand bis(2-benzimidazolyl)ethane (abbreviated as dbz) having the general formula [Cu(dbz)Cl2](Hb) x . (in which Hb = EtOH or MeOH and x = 1/2 or 1). The X-ray crystal structure has been solved from four different compounds, the green α-[CuCl2dbz](C2H5OH)1/2 (1) [27], the red β-[CuCl2dbz](C2H5OH)1/2 (2), the red α-[CuCl2dbz](CH3OH) (3) and the blue–green compound β-[CuCl2dbz](CH3OH) (4). The characterization was further done by spectroscopic methods and EPR. This study is a rare case of distortion isomerism in which four different CuCl2 compounds with the same ligand is fully characterized, also by X-ray crystallography.
Experimental section
General
C, H, N determinations were performed on a Perkin Elmer 2400 Series II analyzer. Ligand field spectra were obtained on a Perkin-Elmer Lambda 900 spectrophotometer, using the diffuse reflectance technique, with MgO as a reference. X-band powder EPR spectra were obtained on a Jeol RE2x electron spin resonance spectrometer using DPPH (g = 2.0036) as a standard. FTIR spectra were obtained on a Perkin Elmer Paragon 1000 FTIR spectrophotometer equipped with a Golden Gate ATR device, using the reflectance technique (4,000–300 cm−1, res. 4 cm−1).
Synthesis of the coordination compounds
The compounds were prepared according to the following general procedure: 1.2 mmol of CuCl2 and 1.2 mmol of the ligand dbz were each dissolved in 10 ml of methanol or ethanol. The Cu(II) salt solution was then added slowly to the ligand solution and filtered to remove any undissolved material. Usually after a few days the products separated as a mixture of red and green crystals for the ethanol solution or red and blue–green crystals for the methanol solution. The different coloured crystals were separated manually for spectroscopic measurements and X-ray crystallography.
Changing the Cu/ligand ratio appeared to have a small effect on the crystallisation of the different coloured products, but it was not possible to synthesize these isomers completely separately. Satisfactory elemental (C, H, N) analysis were obtained.
Crystal structure determination and refinement
A crystal was selected and mounted to a glass fiber using the oil-drop method; data were collected on a Nonius KappaCCD diffractometer (graphite-monochromated Mo-Kα radiation). The intensity data were corrected for Lorentz and polarization effects, for absorption and for extinction. The structures were solved by direct methods. The programs COLLECT [28], SHELXS-97 [29], SHELXL-97 [30] were used for data reduction, structure solution and structure refinement, respectively. Refinement of F 2 was done against all reflections. All non-hydrogen atoms were refined anisotropically. The ethanol molecule and the Cl atoms coordinated to the Cu1A atom in compound (2) were found in disordered positions. These disordered atoms were refined in the two positions with population parameter 0.5. All H atoms were introduced in calculated positions and refined with fixed geometry with respect to their carrier atoms. Crystallographic data are presented in Table 1
Results and discussion
Description of the crystal structures
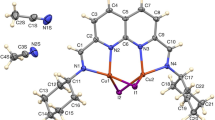
A thermal ellipsoid plot, together with the numbering scheme for the four compounds is shown in Figs. 1–4, for compounds 1 to 4, respectively. The selected bond lengths and bond angles are given in Table 2.
The structure of compounds 1 and 2 contains two chemically almost identical, but crystallographically independent, CuCl2-dbz units and one non-coordinating ethanol molecule, which is present in a disordered position for compound 2. In compound 2 also one of the two units contains the two chloride atoms in disordered positions, which appears to be related to the hydrogen-bonding interaction with the disordered ethanol. Compounds 3 and 4 have only one crystallographic unit and one non-coordinating methanol molecule.
Each copper(II) atom is tetrahedrally surrounded by two nitrogens of the chelating bidentate ligand and two chloride anions. The Cu–N distances vary from 1.801(5) to 2.141(4) Å and the Cu–Cl distances vary from 2.152(3) to 2.279(2) Å (the disordered Cu–Cl distances are not considered), which are normal distances for CuCl2 tetrahedral compounds. The smallest tetrahedral angles (N–Cu–N) of the 7-membered chelate ring differs from 89.12(18)° to 99.8(2)° in the four compounds. Structural comparison of the four compounds is given in Table 3. The fact that in compound 2 the Cl atoms in one of the units is disordered makes it somewhat difficult to compare these compounds with each other.
One of the important differences is the dihedral angle between the Cl–Cu–Cl and the N–Cu–N planes in the four compounds (see Table 3). These tetrahedral angles differ significantly from the tetrahedral angles reported for the copper(II) halide compounds with tbz and methylated tbz (dihedral angles 62.52 and 65.43°); values of 90° would be expected for undistorted tetrahedral geometry, so the present compounds are more close to tetrahedral than the tbz or methylated tbz compounds, perhaps also influenced by the fact that tbz and methylated tbz compounds forms an eight-membered chelate ring [19], while the present compounds forms a seven-membered chelate ring.
The lattice structure is additionally stabilized by stacking between the benzimidazole groups of different units (Ring–Ring distances are 3.552, 3.416, 3.500, 3.451 Å, for compounds 1 to 4, respectively). An extensive hydrogen bond system is present between the N–H of the ligand and the chloride ions; between the N–H and the oxygen atom of the alcohol molecule and between the oxygen atom of the alcohol molecule and the chloride ions. Details of the H bonds are given in Table 2. This great variety in H bonds is likely to be responsible for the different distortion isomers found.
Ligand field and infrared spectroscopy
The diffuse reflectance spectra of the powdered solids are given in Table 4, together with the EPR values. The compounds 1 to 3 show the typical tetrahedral-type ligand field spectrum of a very broad (split) band with a centre around 12.0−9.7 × 103 cm−1, whereas compound 4 has a very broad band centred around 15.0 × 103 cm−1, which are known to be in the common range of absorptions for (square-planar distorted) tetrahedral copper(II) compounds [14, 19, 31].
The infrared spectra of the four compounds show only some differences in the characteristic benzimidazole C–H bending vibrations in the 700–800 cm−1 area. The benzimidazole-CH bending vibrations between 750–720 cm−1 are known to be very sensitive to structure differences and orientations of the benzene part of the molecule in the lattice [32]. In the dbz free ligand the imidazole bending vibration is observed at 775 cm−1 and the benzimidazole CH-out-of-plane bending at 750 and 739 cm−1 [32a].
These infrared vibrations in compound 1 are observed at 761, 753, 750, 744 and 736 cm−1; the two red compounds, 2 and 3 have almost the same pattern, bands at 761, 753, 746, 733 cm−1 (compound 2) and 762, 753, 747, 733 cm−1 (for compound 3). The blue–green compound 4 has a pattern of three vibrations at 764, 757 and 752 cm−1. The ligand N–H stretches vary for each of the compounds (3,464–3,000 cm−1), which is related to their hydrogen bonding situation [34]. The bands are, however, too broad to allow a study with comparision of H-bond distances, also because of the lattice EtOH and MeOH OH-stretching vibrations, which occur in the same region.
EPR
X-band EPR spectra of polycrystalline powders of the complexes were recorded at room temperature and 77 K. The resolution did not improve very much upon cooling to 77 K. As shown in Table 2 the compounds show an axial S = 1/2 signal with a g ⊥ at 2.11–2.07 and an unresolved g // value in the range of 2.23–2.32. EPR measurements as frozen solutions (DMSO/MeOH mixtures) resulted in resolved spectra with g ⊥ of 2.07, g // 2.32 and A// between 12.7–14.4. These values differ from the earlier reported CuCl2-(tbz) compounds which show in similar solutions solution a trigonal-bipyramidal geometry; perhaps some dmso is coordinating to the metal of the CuCl2-(tbz), thereby changing the spectral parameters [14]. The g values obtained are nevertheless in the usual range for copper in a (distorted) tetrahedral environment [14, 16, 19, 21].
Conclusion
The present study has provided four new samples of Cu(II) distortion isomeric compounds with the ligand dbz. So far only a few examples of CuCl2-based distortion isomers are known which have been fully characterized by X-ray analysis [14, 26d, h], and the present species are the first examples in which four different compounds of the same ligand and metal salt and the same chromophore are fully characterized by X-ray crystallography. All compounds exhibit a more or less distorted tetrahedral environment and a seven-membered chelate ring is formed. The tetrahedral angles (N–Cu–N) of the chelate rings are close to the calculated value (101.3°) for 7-membered chelate rings [33].
The difference in the dihedral angles for the various isomers can be understood on the basis of different hydrogen bonding due to the fact of two different solvents and differences in packing due to ring–ring stacking, thereby illustrating that the energy differences are small. Another interesting fact is that in compound (2) the chloride anions are disordered, the reason for that is still unknown.
Supplementary material
Crystallographic data (excluding structure factors) for the structures reported in this paper have been deposited with the Cambridge Crystallographic Data Centre as supplementary publications no. CCDC: 602249, 602250, 602251 and 602252, for compounds 1 to 4, respectively. Copies of the data can be obtained free of charge on application to The Director, CCDC, 12 Union Road, Cambridge CB2 1EZ, UK (fax:int code +44(1223)336-033, E-mail: deposit@ccdc.cam.ac.uk).
References
Reedijk J (1987) In: Wilkinson G, Gillard RD, McCleverty JA (eds) Comprehensive coordination chemistry, Pergamon Press, 2:73
Bouwman E, Driessen WL, Reedijk J (1990) Coord Chem Rev 104:143
Sorrell TN (1989) Tetrahedron 45:3
Zanello P, Tamburini S, Vigato PA, Mazzocchin GA (1987) Coord Chem Rev 77:165
Çakir B, Büyükbingöl E, Uçucu Ü, Abbasoglu U, Noyanalpan N (1988) J Fac Pharm Gazi 5:71
Roderick WR, Nordeen CW Jr, Esch AM, Appell von RN (1972) J Med Chem 15:655
Hoorn HJ, Joode P de, Driessen WL, Reedijk J (1995) React Funct Polym 27:223
Berkel PM van, Dijkstra DJ, Driessen WL, Reedijk J, Sherrington DC (1995) React Funct Polym 28:39
Piquet C, Bünzli J-CG, Bernardinelli G, Hopfgartner G, Williams AF (1995) J Alloys Comp 225:324
Bernardinelli G, Kübel-Pollak A, Rüttimann S, Williams AF (1992) Chimia 46:155
Bernardinelli G, Kübel-Pollak A, Rüttimann S, Williams AF (1993) Zeitschrift Kristallogr 203:132
Albada GA van, Lakin MT, Veldman N, Spek AL, Reedijk J (1995) Inorg Chem 34:4910
Albada GA van, Smeets WJJ, Spek AL, Reedijk J (1997) Inorg Chim Acta 260:151
Albada GA van, Smeets WJJ, Spek AL, Reedijk J (1999) Inorg Chim Acta 288:220
Albada GA van, Smeets WJJ, Veldman N, Spek AL, Reedijk J (1999) Inorg Chim Acta 290:105
Albada GA van, Smeets WJJ, Spek AL, Reedijk J (2000) Inorg Chim Acta 299:35
Albada GA van, Veldman N, Spek AL, Reedijk J (2000) Chem Crystallogr 30:69
Albada GA van, Riggio I, Mutikainen I, Turpeinen U, Reedijk J (2000) Chem Cryst 30:793
Riggio I, Albada GA van, Ellis DD, Mutikainen I, Spek AL, Turpeinen U, Reedijk J, (2001) Polyhedron 20:2659
Albada GA van, Mutikainen I, Riggio I, Turpeinen U, Reedijk J (2002) Polyhedron 21:141
Hathaway BJ (1987) In: Wilkinson G, Gillard RD, McCleverty JA (eds) Comprehensive coordination chemistry,vol 5, Pergamon, Oxford
Gažo J, Bersuker IB, Garaj J, Kabešová M, Kohout J, Langfelderová H, Melník M, Serátor M, Valach F (1976) Coord Chem Rev 19:253
Palaniandavar M, Butcher RJ, Addison AW (1996) Inorg Chem 35:467
Gažo J (1974) Pure Appl Chem 38:279
Melník M (1982) Coord Chem Rev 47:239
(a) Haddad SF, Pickardt J (1993) Transition Met Chem 18:377; (b)Foley J, Tyagi S, Hathaway BJ (1984) J Chem Soc, Dalton Trans 1; (c) Baran P, Koman M, Valigura D, Mroziński J (1991) J Chem Soc, Dalton Trans 1385; (d) Kelly PF, Slawin AMZ, Waring KW (1997) J Chem Soc, Dalton Trans 2853; (e) Pajunen A, Smolander K, Belinskij I (1972) Suomen Kemistilehti B45:317; (f) Glass RS, Sabahi M, Hojjatie M, Wilson GS (1987) Inorg Chem 26:2196; (g) Haanstra WG, Donk WAJW van der, Driessen WL, Reedijk J, Wood JS, Drew MGB (1990) J Chem Soc, Dalton Trans 3123; (h) Langfelderová H, Macásková L, Melník M, Kabešová M, Gažo J (1978) Z Anorg Allg Chem 445:233
The X-ray structure of compound 1 was measured for the first time in 1995, but with a significantly higher R-value (see CSD reference ESIHIX, Spek AL, Veldman N, Albada GAvan, Reedijk J Private Comm) Current data are more accurate
Nonius, 2002 COLLECT Nonius BV, Delft, The Netherlands
Sheldrick, GM SHELXS-97, Program for crystal structure determination, University of Göttingen, Germany, 1997
Sheldrick, GM (1997) SHELXL-97 Program for the Refinement of Crystal Structures University of Göttingen, Germany
Lever ABP (1984) Inorganic electronic spectroscopy, 2nd Ed, Elsevier, Amsterdam
(a) Drolet DP, Manuta DM, Lees AJ, Katnani AD, Coyle GJ (1988) Inorg Chim Acta 146:173; (b) Boinnard D, Cassoux P, Petrouleas V, Savariault J-M, Tuchagues J-P (1990) Inorg Chem 29:4114; (c) Cordes MM, Walter JL (1968) Spectrochim Acta 24A:1421
Broughton V, Bernardinelli G, Williams AF (1998) Inorg Chim Acta 275:276–279
Bellamy LJ, Owen AJ (1969) Spectr Chim Acta 25A:329
Acknowledgements
The work described in the present paper has been supported by the Leiden University Study group WFMO (Werkgroep Fundamenteel Materialen Onderzoek).
Financial support and coordination by the FP6 Network of Excellence “Magmanet”(contract number 515767) is kindly acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
van Albada, G.A., Mutikainen, I., Turpeinen, U. et al. Distortion Isomerism of Cu(II) Chloride Adducts with Bis(2-benzimidazolyl)ethane. Synthesis, Characterization, X-ray Structures and Spectroscopy of Four Different Isomers. J Chem Crystallogr 37, 489–496 (2007). https://doi.org/10.1007/s10870-007-9202-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10870-007-9202-0