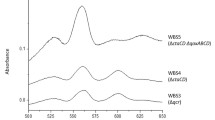
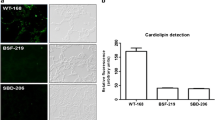
Abstract
Bacillus subtilis is a Gram-positive bacterium with a respiratory chain embedded in the cytoplasmic membrane. The respiratory chain is bifurcated after menaquinol into a cytochrome b6c + caa3 branch and a branch with up to three quinol oxidases. The complexes that generate the proton gradient are b6c, associated with caa3 and aa3 oxidase. The b6c and caa3 complexes form a supercomplex, and it is proposed to form respiratory strings in the membrane. There is still information missing about the quinol branch and if the primary oxidase quinol aa3 is associated with the electron donor complexes. It is unclear whether succinate quinone reductase (SQR) can form associations with the quinol branch or the cytochrome branch. In this paper, we show the separation of an almost pure b6c complex associated with cytochromes c550 and c551. We obtained a b6c + caa3 supercomplex of 600 kDa and SQR, aa3, and NADH dehydrogenase by dodecyl maltoside solubilization and separation of the respiratory chain components by ionic exchange chromatography. We found that aa3 does not associate with other complexes. SQR was associated with the b6c complex in a mutant lacking aa3. This association could facilitate electron transfer from SQR to menaquinone-7. The lack of associations between the abundant quinol oxidase aa3 and other complexes is a feature we cannot explain yet.







Similar content being viewed by others
Data availability statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Acín-Pérez R, Bayona-Bafaluy MP, Fernández-Silva P et al (2004) Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol Cell 13:805–815. https://doi.org/10.1016/S1097-2765(04)00124-8
Andrews D, Mattatall NR, Arnold D, Hill BC (2005) Expression, purification, and characterization of the Cu A-cytochrome c domain from subunit II of the Bacillus subtilis cytochrome caa3 complex in Escherichia coli. Protein Expr Purif 42:227–235. https://doi.org/10.1016/j.pep.2004.11.009
Azarkina N, Siletsky S, Borisov V et al (1999) A cytochrome bb’-type quinol oxidase in Bacillus subtilis strain 168. J Biol Chem 274:32810–32817. https://doi.org/10.1074/jbc.274.46.32810
Barrera-Rojas J, de la Vara LG, Ríos-Castro E et al (2018) The distribution of divinyl chlorophylls a and b and the presence of ferredoxin-NADP + reductase in Prochlorococcus marinus MIT9313 thylakoid membranes. https://doi.org/10.1016/j.heliyon.2018.e01100. Heliyon 4:
Bengtsson J, Rivolta C, Hederstedt L, Karamata D (1999) Bacillus subtilis contains two small c-type cytochromes with homologous heme domains but different types of membrane anchors. J Biol Chem 274:26179–26184. https://doi.org/10.1074/jbc.274.37.26179
Berry EA, Trumpower BL (1985) Isolation of ubiquinol oxidase from Paracoccus denitrificans and resolution into cytochrome bc1 and cytochrome c-aa3 complexes. Journal of Biological Chemistry 260:2458–2467. https://doi.org/10.1016/s0021-9258(18)89576-x
de Vrij W, Konings WN (1987) Kinetic characterization of cytochrome c oxidase from Bacillus subtilis. Eur J Biochem / FEBS 166:581–587. https://doi.org/10.1111/j.1432-1033.1987.tb13553.x
Deutsch EW, Mendoza L, Shteynberg D et al (2010) A guided tour of the Trans-Proteomic Pipeline. Proteomics 10:1150–1159. https://doi.org/10.1002/PMIC.200900375
Diaz F, Garcia S, Padgett KR, Moraes CT (2012) A defect in the mitochondrial complex III, but not complex IV, triggers early ROS-dependent damage in defined brain regions. Hum Mol Genet 21:5066–5077. https://doi.org/10.1093/hmg/dds350
Fedotovskaya O, Albertsson I, Nordlund G et al (2021) Identification of a cytochrome bc1-aa3 supercomplex in Rhodobacter sphaeroides. Biochim Biophys Acta Bioenerg 1862. https://doi.org/10.1016/j.bbabio.2021.148433
García Montes De Oca LYJ, Chagolla-López A, De La Vara LG et al (2012) The composition of the Bacillus subtilis aerobic respiratory chain supercomplexes. J Bioenerg Biomembr 44:473–486. https://doi.org/10.1007/s10863-012-9454-z
García Montes de Oca LYJ, Cabellos Avelar T, Picón Garrido GI et al (2016) Cardiolipin deficiency causes a dissociation of the b6c:caa3 megacomplex in B. subtilis membranes. J Bioenerg Biomembr 48:451–467. https://doi.org/10.1007/s10863-016-9671-y
Genova ML, Lenaz G (2014) Functional role of mitochondrial respiratory supercomplexes. Biochimica et Biophysica Acta (BBA). - Bioenergetics 1837:427–443. https://doi.org/10.1016/j.bbabio.2013.11.002
Guo R, Gu J, Zong S et al (2018) Structure and mechanism of mitochondrial electron transport chain. Biomed J 41:9–20. https://doi.org/10.1016/j.bj.2017.12.001
Gyan S, Shiohira Y, Sato I et al (2006) Regulatory loop between redox sensing of the NADH/NAD + ratio by Rex (YdiH) and oxidation of NADH by NADH dehydrogenase Ndh in Bacillus subtilis. J Bacteriol 188:7062–7071. https://doi.org/10.1128/JB.00601-06
Hägerhäll C, von Wachenfeldt C, Hederstedt L, Aasa R (1992) Two Hemes in Bacillus subtilis Succinate: Menaquinone Oxidoreductase (Complex II). Biochemistry 31:7411–7421. https://doi.org/10.1021/bi00147a028
Hederstedt L (1980) Cytochrome b reducible by succinate in an isolated succinate dehydrogenase-cytochrome b complex from Bacillus subtilis membranes. J Bacteriol 144:933–940. https://doi.org/10.1128/JB.144.3.933-940.1980
Hederstedt L (2021) Molecular Biology of Bacillus subtilis Cytochromes anno 2020. Biochem (Moscow) 86:8–21. https://doi.org/10.1134/s0006297921010028
Hederstedt L (1986) [38] Molecular properties, genetics, and biosynthesis of Bacillus subtilis succinate dehydrogenase complex. In: Methods in Enzymology. pp 399–414. https://doi.org/10.1016/S0076-6879(86)26040-1
Henning W, Vo L, Albanese J, Hill BC (1995) High yield purification of cytochrome aa3 and cytochrome caa3 oxidases from Bacillus subtilis plasma membranes. Biochem J 309:279–283. https://doi.org/10.1042/bj3090279
Konings WN, Freese E (1972) Amino acid transport in membrane vesicles of Bacillus subtilis. J Biol Chem 247:2408–2418. https://doi.org/10.1016/s0021-9258(19)45444-6
Kovács Á (2019) Bacillus subtilis. Trends Microbiol 27:724–725. https://doi.org/10.1016/j.tim.2019.03.008
Kunst F, Ogasawara N, Moszer I et al (1997) The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249–256. https://doi.org/10.1038/36786
Kutoh E, Sone N (1988) Quinol-cytochrome c oxidoreductase from the thermophilic bacterium PS3. Purification and properties of a cytochrome bc1(b6f) complex. J Biol Chem 263:9020–9026
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. https://doi.org/10.1038/227680a0
Lauraeus M, Haltia T, Saraste M, Wikstrom M (1991) Bacillus subtilis Expresses Two Kinds of Haem-A- Containing Terminal Oxidases. EurJBiochem 197:699–705. https://doi.org/10.1111/j.1432-1033.1991.tb15961.x
Lauraeus M, Wikström M (1993) The terminal quinol oxidases of Bacillus subtilis have different energy conservation properties. J Biol Chem 268:11470–11473
Ljungdahl PO, Pennoyer JD, Robertson DE, Trumpower BL (1987) Purification of highly active cytochrome bc1 complexes from phylogenetically diverse species by a single chromatographic procedure. BBA - Bioenergetics 891:227–241. https://doi.org/10.1016/0005-2728(87)90218-0
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275.
Magalon A, Alberge F (2016) Distribution and dynamics of OXPHOS complexes in the bacterial cytoplasmic membrane. Biochim Biophys Acta-Bioenergetics 1857:198-213. https://doi.org/10.1016/j.bbabio.2015.10.015
Markwell MAK, Haas SM, Bieber LL, Tolbert NE (1978) A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem 87:206–210. https://doi.org/10.1016/0003-2697(78)90586-9
Matsson M, Tolstoy D, Aasa R, Hederstedt L (2000) The distal heme center in Bacillus subtilis succinate:quinone reductase is crucial for electron transfer to menaquinone. Biochemistry 39:8617–8624. https://doi.org/10.1021/bi000271m
Melo AMP, Teixeira M (2016) Supramolecular organization of bacterial aerobic respiratory chains: From cells and back. Biochim Biophys Acta 1857:190–197. https://doi.org/10.1016/j.bbabio.2015.11.001
Moszer I, Jones LM, Moreira S et al (2002) SubtiList: The reference database for the Bacillus subtilis genome. Nucleic Acids Res 30:62–65. https://doi.org/10.1093/nar/30.1.62
Nicholls DG, Ferguson S (2013) Bioenergetics: Fourth Edition. Bioenergetics: Fourth Edition 1–419. https://doi.org/10.1016/C2010-0-64902-9
Nubel E, Wittig I, Kerscher S et al (2009) Two-dimensional native electrophoretic analysis of respiratory supercomplexes from Yarrowia lipolytica. Proteomics 9:2408–2418. https://doi.org/10.1002/pmic.200800632
Papa S, Capitanio N, Glaser P, Villani G (1994) The proton pump of heme-copper oxidases. Cell Biol Int 18:345–356. https://doi.org/10.1006/CBIR.1994.1084
Rajendran C, Ermler U, Ludwig B, Michel H (2010) Structure at 1.5 Å resolution of cytochrome c552 with its flexible linker segment, a membrane-anchored protein from Paracoccus denitrificans. Acta Crystallogr D Biol Crystallogr 66:850–854. https://doi.org/10.1107/S0907444910019396
Sabar M, Balk J, Leaver CJ (2005) Histochemical staining and quantification of plant mitochondrial respiratory chain complexes using blue-native polyacrylamide gel electrophoresis. Plant J 44:893–901. https://doi.org/10.1111/j.1365-313X.2005.02577.x
Sakamoto J, Koga E, Mizuta T et al (1999) Gene structure and quinol oxidase activity of a cytochrome bd-type oxidase from Bacillus stearothermophilus. Biochim Biophys Acta Bioenerg 1411:147–158. https://doi.org/10.1016/S0005-2728(99)00012-2
Santana M, Kunst F, Hullo MF et al (1992) Molecular cloning, sequencing, and physiological characterization of the qox operon from Bacillus subtilis encoding the aa3-600 quinol oxidase. J Biol Chem 267:10225–10231
Saraste M, Metso T, Nakari T et al (1991) The Bacillus subtilis cytochrome-c oxidase. Variations on a conserved protein theme. Eur J Biochem / FEBS 195:517–525. https://doi.org/10.1007/s12275-008-0045-y
Schaägger H, Cramer WA, von Jagow G (1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem 217:220–230. https://doi.org/10.1006/abio.1994.1112
Schirawski J, Unden G (1998) Menaquinone-dependent succinate dehydrogenase of bacteria catalyzes reversed electron transport driven by the proton potential. Eur J Biochem 257:210–215. https://doi.org/10.1046/j.1432-1327.1998.2570210.x
Schnorpfeil M, Janausch IG, Biel S et al (2001) Generation of a proton potential by succinate dehydrogenase of Bacillus subtilis functioning as a fumarate reductase. Eur J Biochem 268:3069–3074. https://doi.org/10.1046/j.1432-1327.2001.02202.x
Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal Chem 68:850–858. https://doi.org/10.1021/ac950914h
Singer TP (1979) Mitochondrial electron-transport inhibitors. Methods Enzymol 55:454–62. https://doi.org/10.1016/0076-6879(79)55059-9
Sonenshein AL, Hoch JA, Losick R (2001) Bacillus subtilis and Its Closest Relatives. American Society of Microbiology. https://doi.org/10.1128/9781555817992
Sousa PMF, Silva STN, Hood BL et al (2011) Supramolecular organizations in the aerobic respiratory chain of Escherichia coli. Biochimie 93:418–425. https://doi.org/10.1016/J.BIOCHI.2010.10.014
Sousa PMF, Videira M, Santos M, a F et al (2013) The bc:caa3 supercomplexes from the Gram-positive bacterium Bacillus subtilis respiratory chain: A megacomplex organization? Arch Biochem Biophys 537:153–160. https://doi.org/10.1016/j.abb.2013.07.012
Winstedt L, von Wachenfeldt C (2000) Terminal oxidases of Bacillus subtilis strain 168: one quinol oxidase, cytochrome aa(3) or cytochrome bd, is required for aerobic growth. J Bacteriol 182:6557–6564. https://doi.org/10.1128/JB.182.23.6557-6564.2000
Winstedt L, Yoshida KI, Fujita Y, Von Wachenfeldt C (1998) Cytochrome bd biosynthesis in Bacillus subtilis: Characterization of the cydABCD operon. J Bacteriol 180:6571–6580. https://doi.org/10.1128/JB.180.24.6571-6580.1998
Wittig I, Beckhaus T, Wumaier Z et al (2010) Mass estimation of native proteins by blue native electrophoresis: Principles and practical hints. Mol Cell Proteomics 9:2149–2161. https://doi.org/10.1074/mcp.M900526-MCP200
Wittig I, Karas M, Schägger H (2007) High Resolution Clear Native Electrophoresis for In-gel Functional Assays and Fluorescence Studies of Membrane Protein Complexes. Mol Cell Proteom 6:1215–1225. https://doi.org/10.1074/mcp.M700076-MCP200
Yu J, Hederstedt L, Piggot PJ (1995) The cytochrome bc complex (menaquinone:cytochrome c reductase) in Bacillus subtilis has a nontraditional subunit organization. J Bacteriol 177:6751–6760. https://doi.org/10.1128/jb.177.23.6751-6760.1995
Yu J, le Brun NE (1998) Studies of the cytochrome subunits of menaquinone:cytochrome c reductase (bc complex) of Bacillus subtilis. Evidence for the covalent attachment of heme to the cytochrome b subunit. J Biol Chem 273:8860–8866. https://doi.org/10.1074/jbc.273.15.8860
Acknowledgements
This work is the requirement to obtain the PhD degree in Sciences (Experimental Biology) for the student Gerardo Ignacio Picón Garrido in the Posgrado en Ciencias Biológicas, U.N.A.M. The work was supported by Grants DGAPA, PAPIIT IN219115 and IN219118. We are grateful to Biologist Bertha Pérez Gómez and Dr. Jesús Barrera Rojas for their technical assistance.
Funding
Funding for this research was obtained from Dirección General de Asuntos del Personal Académico (D.G.A.P.A.) (Universidad Nacional Autónoma de México, U.N.A.M.). Research Grants: IN219115 and IN912118.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design.
GIPG: he grew Bacillus subtilis WT and mutants, prepared membranes for mutants, obtained the solubilization and isolation of respiratory complexes. Prepared the CNE and 2D SDS PAGE. APGG: also grew B. subtilis WT and assisted during the chromatographies, obtained the fractions and the visible spectroscopy analysis. AGL: prepared the samples and obtained the mass spectrometry analysis from the samples of CNE. LGV: contributed to the analysis of the results from mass spectrometry, selected the best ID values for all the proteins and assisted in the verification and analysis of all the mass spectrometry samples. CGL: donated material and chemicals, contributed to the discussion of the results and writing of the manuscript. EBGC: obtained funding and proposed the experiments, assisted in the analysis of results and discussion of the results and writing the manuscript.
Corresponding author
Ethics declarations
Competing interests statement
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.

10863_2022_9951_MOESM1_ESM.png
Supplementary Material 1: Spectra of pools I-III obtained from the chromatography of solubilized membranes of WT (letters A through C) and solubilized membranes from mutant ∆Qox (LUH46) (letters D through F). Discontinuous line: ascorbate reduced membranes; continuous line: dithionite reduced membranes; dash-dotted line: the difference spectrum obtained by subtracting the dithionite-reduced minus ascorbate-added spectra.

10863_2022_9951_MOESM2_ESM.png
Supplementary Material 2: Spectra of membranes isolated from the mutants used in this work compared to the WT spectrum. A: dithionite-added spectrum of ∆Qox-aa3 membranes (dotted line) compared to the WT spectrum (continuous line). B: dithionite-added spectrum of ∆SQR membranes (dotted line) compared to the WT spectrum (continuous line). C: dithionite-added spectrum of ∆b6c membranes (dotted line) compared to the WT spectrum (continuous line)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Picón Garrido, G.I., García García, A.P., González de la Vara, L. et al. Separation and analysis of Bacillus subtilis respiratory chain complexes. J Bioenerg Biomembr 54, 251–271 (2022). https://doi.org/10.1007/s10863-022-09951-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10863-022-09951-6