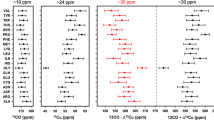
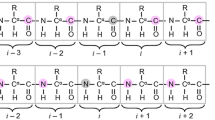
Abstract
The solution NMR resonance assignment of the protein backbone is most commonly carried out using triple resonance experiments that involve 15N and 1HN resonances. The assignment becomes problematic when there is resonance overlap of 15N–1HN cross peaks. For such residues, one cannot unambiguously link the “left” side of the NH root to the “right” side, and the residues associated with such overlapping HN resonances remain often unassigned. Here we present a solution to this problem: a hybrid (4d,3d) reduced-dimensionality HN(CO)CA(CON)CA sequence. In this experiment, the Ca(i) resonance is modulated with the frequency of the Ca(i−1) resonance, which helps in resolving the ambiguity involved in connecting the Ca(i) and Ca(i−1) resonances for overlapping NH roots. The experiment has limited sensitivity, and is only suited for small or unfolded proteins. In a companion experiment, (4d,3d) reduced-dimensionality HNCO(N)CA, the Ca(i) resonance is modulated with the frequency of the CO(i−1) resonance, hence resolving the ambiguity existent in pairing up the Ca(i) and CO(i−1) resonances for overlapping NH roots.




Similar content being viewed by others
References
Atreya HS, Szyperski T (2004) G-matrix Fourier transform NMR spectroscopy for complete protein resonance assignment. Proc Natl Acad Sci U S A 101(26):9642–9647
Cavanagh J, Fairbrother W, Palmer AG III, Rance M, Skelton NJ (2007) Protein NMR spectroscopy: principles and practice, 2nd edn. Elsevier Academic Press, Amsterdam
Crippen GM, Rousaki A, Revington M, Zhang Y, Zuiderweg ER (2010) Saga: rapid automatic mainchain NMR assignment for large proteins. J Biomol NMR 46(4):281–298
Fiorito F, Hiller S, Wider G, Wuthrich K (2006) Automated resonance assignment of proteins: 6d apsy-NMR. J Biomol NMR 35(1):27–37
Hiller S, Fiorito F, Wuthrich K, Wider G (2005) Automated projection spectroscopy (apsy). Proc Natl Acad Sci USA 102(31):10876–10881
Hiller S, Wasmer C, Wider G, Wuthrich K (2007) Sequence-specific resonance assignment of soluble nonglobular proteins by 7d apsy-NMR spectroscopy. J Am Chem Soc 129(35):10823–10828
Kupce E, Freeman R (2004) Projection-reconstruction technique for speeding up multidimensional NMR spectroscopy. J Am Chem Soc 126(20):6429–6440
Kupce E, Freeman R (2006) Hyperdimensional NMR spectroscopy. J Am Chem Soc 128(18):6020–6021
Loria JP, Rance M, Palmer AG III (1999) Transverse-relaxation-optimized (trosy) gradient-enhanced triple-resonance NMR spectroscopy. J Magn Reson 141(1):180–184
Moseley HN, Monleon D, Montelione GT (2001) Automatic determination of protein backbone resonance assignments from triple resonance nuclear magnetic resonance data. Methods Enzymol 339:91–108
Rance M, Loria JP, AGr Palmer (1999) Sensitivity improvement of transverse relaxation-optimized spectroscopy. J Magn Reson 136(1):92–101
Salzmann M, Pervushin K, Wider G, Senn H, Wuthrich K (1998) Trosy in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc Natl Acad Sci U S A 95(23):13585–13590
Schulte-Herbruggen T, Sorensen OW (2000) Clean trosy: compensation for relaxation-induced artifacts. J Magn Reson 144(1):123–128
Shen Y, Atreya HS, Liu G, Szyperski T (2005) G-matrix fourier transform noesy-based protocol for high-quality protein structure determination. J Am Chem Soc 127(25):9085–9099
Simorre JP, Brutscher B, Caffrey MS, Marion D (1994) Assignment of NMR spectra of proteins using triple-resonance two-dimensional experiments. J Biomol NMR 4(3):325–333
Szyperski T, Wider G, Bushweller J, Wuthrich K (1993) Reduced dimensionality in triple-resonance NMR experiments. J Am Chem Soc 115:9307–9308
Yi L, Jenkins PM, Leichert LI, Jakob U, Martens JR, Ragsdale SW (2009) Heme regulatory motifs in heme oxygenase-2 form a thiol/disulfide redox switch that responds to the cellular redox state. J Biol Chem 284(31):20556–20561
Acknowledgments
We acknowledge NIH R01HL 102662; ERPZ also acknowledges NIH ARRA GM063027-S2.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bagai, I., Ragsdale, S.W. & Zuiderweg, E.R.P. Pseudo-4D triple resonance experiments to resolve HN overlap in the backbone assignment of unfolded proteins. J Biomol NMR 49, 69–74 (2011). https://doi.org/10.1007/s10858-010-9465-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-010-9465-1