Abstract
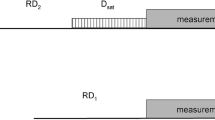
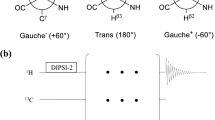
A novel NMR pulse sequence is introduced to determine the glycosidic torsion angle χ in 13C,15N-labeled oligonucleotides. The quantitative Γ-HCNCH measures the dipolar cross-correlated relaxation rates \(\Gamma_{{\rm C6H6,C1}^{\prime}{\rm H1}^{\prime}}^{\rm DD,DD}\) (pyrimidines) and \(\Gamma_{{\rm C8H8,C1}^{\prime}{\rm H1}^{\prime}}^{\rm DD,DD}\) (purines). Cross-correlated relaxation rates of a 13C,15N-labeled RNA 14mer containing a cUUCGg tetraloop were determined and yielded χ-angles that agreed remarkably well with data derived from the X-ray structure of the tetraloop. In addition, the method was applied to the larger stemloop D (SLD) subdomain of the Coxsackievirus B3 cloverleaf 30mer RNA and the effect of anisotropic rotational motion was examined for this molecule. It could be shown that the χ-angle determination especially for nucleotides in the anti conformation was very accurate and the method was ideally suited to distinguish between the syn and the anti-conformation of all four types of nucleotides.










Similar content being viewed by others
References
Allain FH, Varani G (1995) Structure of the P1 helix from group I self-splicing introns. J Mol Biol 250:333–353
Ban N, Nissen P, Hansen J, Moore PB, Steitz TA (2000) The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289:905–920
Banci L, Bertini I, Felli IC, Hajieva P, Viezzoli MS (2001) Side chain mobility as monitored by CH–CH cross correlation: the example of cytochrome b5. J Biomol NMR 20:1–10
Batey RT, Inada M, Kujawinski E, Puglisi JD, Williamson JR (1992) Preparation of isotopically labeled ribonucleotides for multidimensional NMR spectroscopy of RNA. Nucleic Acids Res 20:4515–4523
Batey RT, Battiste JL, Williamson JR (1995) Preparation of isotopically enriched RNAs for heteronuclear NMR. Methods Enzymol 261:300–322
Boisbouvier J, Bax A (2002) Long-range magnetization transfer between uncoupled nuclei by dipole–dipole cross-correlated relaxation: a precise probe of beta-sheet geometry in proteins. J Am Chem Soc 124:11038–11045
Carlomagno T, Felli IC, Czech M, Fischer R, Sprinzl M, Griesinger C (1999) Transferred cross-correlated relaxation: application to the determination of sugar pucker in an aminoacylated tRNA-mimetic weakly bound to EF-Tu. J Am Chem Soc 121:1945–1948
Carlomagno T, Blommers MJ, Meiler J, Cuenoud B, Griesinger C (2001) Determination of aliphatic side-chain conformation using cross-correlated relaxation: application to an extraordinarily stable 2′-aminoethoxy-modified oligonucleotide triplex. J Am Chem Soc 123:7364–7370
Cromsigt J, van Buuren B, Schleucher J, Wijmenga S (2001) Resonance assignment and structure determination for RNA. Methods Enzymol 338:371–399
Duchardt E, Schwalbe H (2005) Residue specific ribose and nucleobase dynamics of the cUUCGg RNA tetraloop motif by MNMR 13C relaxation. J Biomol NMR 32:295–308
Duchardt E, Richter C, Ohlenschläger O, Görlach M, Wöhnert J, Schwalbe H (2004) Determination of the glycosidic bond angle chi in RNA from cross-correlated relaxation of CH dipolar coupling and N chemical shift anisotropy. J Am Chem Soc 126: 1962–1970
Emsley L, Bodenhausen G (1992) Optimization of shaped selective pulses for NMR using a quaternion description of their overall propagators. J Magn Reson 97:135–148
Ennifar E, Nikulin A, Tishchenko S, Serganov A, Nevskaya N, Garber M, Ehresmann B, Ehresmann C, Nikonov S, Dumas P (2000) The crystal structure of UUCG tetraloop. J Mol Biol 304:35–42
Feller SE, Zhang Y, Pastor RW, Brooks BR (1995) Constant pressure molecular dynamics simulation: the Langevin piston method. J Chem Phys 103:4613–4621
Felli I, Richter C, Griesinger C, Schwalbe H (1999) Determination of RNA sugar pucker mode from cross-correlated relaxation in solution NMR spectroscopy. J Am Chem Soc 121:1956–1957
Ferner J, Duchardt E, Villa A, Stock G, Schwalbe H (to be published)
Fiala R, Jiang F, Sklenář V (1998) Sensitivity optimized HCN and HCNCH experiments for 13C/15N labeled oligonucleotides. J Biomol NMR 12:373–383
Fiala R, Munzarova ML, Sklenar V (2004) Experiments for correlating quaternary carbons in RNA bases. J Biomol NMR 29:477–490
Foloppe N, MacKerell AD Jr (2000) All-atom empirical force field for nucleic acids: I. Parameter optimization based on small molecule and condensed phase macromolecular target data. J Comp Chem 21:86–104
Fürtig B, Richter C, Wöhnert J, Schwalbe H (2003) NMR-spectroscopy of RNA. ChemBioChem 4:936–962
Fürtig B, Richter C, Bermel W, Schwalbe H (2004) New NMR experiments for RNA nucleobase resonance assignment and chemical shift analysis of an RNA UUCG tetraloop. J Biomol NMR 28:69–79
Garcia de la Torre J, Huertas ML, Carrasco B (2000) HYDRONMR: prediction of NMR relaxation of globular proteins from atomic-level structures and hydrodynamic calculations. J Magn Reson 147:138–146
Geen HF, Freeman R (1991) Band-selective radiofrequency pulses. J Magn Reson 93:93–141
Hubbard PS (1969) Nonexponential relaxation of three-spin systems in nonspherical molecules. J Chem Phys 51:1647
Ilin S, Bosques C, Turner C, Schwalbe H (2003) Gamma-HMBC: an NMR experiment for the conformational analysis of the o-glycosidic linkage in glycopeptides. Angew Chem Int Ed 42:1394–1397
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79: 926–935
Kay L, Keifer P, Saarinen T (1992) Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J Am Chem Soc 114:10663–10665
Kloiber K, Schuler W, Konrat R (2002) Automated NMR determination of protein backbone dihedral angles from cross-correlated spin relaxation. J Biomol NMR 22:349–363
MacKerell Jr AD, Banavali NK (2000) All-atom empirical force field for nucleic acids: II. Application to molecular dynamics simulations of DNA and RNA in solution. J Comp Chem 21:105–120
Marion D, Ikura M, Tschudin R, Bax AJ (1989) Rapid recording of 2D NMR spectra without phase cycling. Application to the study of hydrogen exchange in proteins. J Magn Reson 85:393–399
Markwick PR, Sprangers R, Sattler M (2005) Local structure and anisotropic backbone dynamics from cross-correlated NMR relaxation in proteins. Angew Chem Int Ed 44:3232–3237
Millet O, Chiarparin E, Pelupessy P, Pons M, Bodenhausen G (1999) Measurement of relaxation rates of NH and Ha backbone protons in proteins with tailored initial conditions. J Magn Reson 139:434–438
Munzarova ML, Sklenar V (2003) DFT analysis of NMR scalar interactions across the glycosidic bond in DNA. J Am Chem Soc 125:3649–3658
Nikonowicz EP, Sirr A, Legault P, Jucker FM, Baer LM, Pardi A (1992) Preparation of 13C and 15N labelled RNAs for heteronuclear multi-dimensional NMR studies. Nucleic Acids Res 20:4507–4513
Ohlenschläger O, Wöhnert J, Bucci E, Seitz S, Hafner S, Ramachandran R, Zell R, Görlach M (2004) The structure of the stemloop D subdomain of coxsackievirus B3 cloverleaf RNA and its interaction with the proteinase 3C. Structure 12:237–248
Pelupessy P, Chiarparin E, Ghose R, Bodenhausen G (1999) Efficient determination of angles subtended by C(alpha)–H(alpha) and N–H(N) vectors in proteins via dipole–dipole cross-correlation. J Biomol NMR 13:375–380
Quant S, Wechselberger R, Wolter M, Wörner K-H, Schell P, Engels J, Griesinger C, Schwalbe H (1994) Chemical synthesis of 13C-labelled monomers for solid-phase and template controlled enzymatic synthesis of DNA and RNA oligomers. Tetrahedron Lett 35:6649–6652
Ravindranathan S, Kim CH, Bodenhausen G (2003) Cross correlations between 13C–1H dipolar interactions and 15N chemical shift anisotropy in nucleic acids. J Biomol NMR 27:365–375
Reif B, Hennig M, Griesinger C (1997) Direct measurement of angles between bond vectors in high-resolution NMR. Science 276:1230–1233
Richter C, Griesinger C, Felli I, Cole PT, Varani G, Schwalbe H (1999) Determination of sugar conformation in large RNA oligonucleotides from analysis of dipole–dipole cross correlated relaxation by solution NMR spectroscopy. J Biomol NMR 15:241–250
Richter C, Reif B, Griesinger C, Schwalbe H (2000) NMR spectroscopic determination of angles α and ζ in RNA from CH–dipolar coupling, P-CSA cross-correlated relaxation. J Am Chem Soc 122:12728–12731
Roehrl MH, Heffron GJ, Wagner G (2005) Correspondence between spin-dynamic phases and pulse program phases of NMR spectrometers. J Magn Reson 174:325–330
Schneider H (1964) Z Naturforsch Teil A 19:510
Schwalbe H, Marino JP, King GC, Wechselberger R, Bermel W, Griesinger C (1994) Determination of a complete set of coupling constants in 13C-labeled oligonucleotides. J Biomol NMR 4:631–644
Schwalbe H, Carlomagno T, Hennig M, Junker J, Reif B, Richter C, Griesinger C (2001) Cross-correlated relaxation for measurement of angles between tensorial interactions. Methods Enzymol 338:35–81
Shaka AJ, Barker PB, Freeman RJ (1985) Computer-optimized decoupling scheme for wideband applications and low-level operation. J Magn Reson 64:547–552
Sklenar V, Peterson RD, Rejante MR, Feigon J (1993a) Two- and three-dimensional HCN experiments for correlating base and sugar resonances in 15N,13C-labeled RNA oligonucleotides. J Biomol NMR 3:721–727
Sklenar V, Rejante MR, Peterson RD, Wang E, Feigon J (1993b) Two-dimensional triple-resonance HCNCH experiment for direct correlation of ribose H1′ and base H8, H6 protons in 13C,15N-labeled RNA oligonucleotides. J Am Chem Soc 115:12181–12182
Sklenar V, Dieckmann T, Butcher SE, Feigon J (1998) Optimization of triple-resonance HCN experiments for application to larger RNA oligonucleotides. J Magn Reson 130:119–124
Stueber D, Grant DM (2002) 13C and (15)N chemical shift tensors in adenosine, guanosine dihydrate, 2′-deoxythymidine, and cytidine. J Am Chem Soc 124:10539–10551
Sychrovsky V, Muller N, Schneider B, Smrecki V, Spirko V, Sponer J, Trantirek L (2005) Sugar pucker modulates the cross-correlated relaxation rates across the glycosidic bond in DNA. J Am Chem Soc 127:14663–14667
Trantirek L, Stefl R, Masse JE, Feigon J, Sklenar V (2002) Determination of the glycosidic torsion angles in uniformly 13C-labeled nucleic acids from vicinal coupling constants 3J(C2)/4-H1′ and 3J(C6)/8-H1′. J Biomol NMR 23:1–12
Tugarinov V, Kay LE (2004) 1H,13C-1H,1H dipolar cross-correlated spin relaxation in methyl groups. J Biomol NMR 29:369–376
Varani G, Aboul-ela F, Allain FH-T (1996) NMR investigation of RNA structure. Prog Nucl Magn Reson Spectrosc 29:51–127
Varani G, Tinoco I Jr (1991) RNA structure and NMR spectroscopy. Q Rev Biophys 24:479–532
Vugmeyster L, Pelupessy P, Vugmeister BE, Abergel D, Bodenhausen G (2004) Cross-correlated relaxation in NMR of macromolecules in the presence of fast and slow internal dynamics. CR Phys 5:377–386
Wang T, Frederick KK, Igumenova TI, Wand AJ, Zuiderweg ER (2005) Changes in calmodulin main-chain dynamics upon ligand binding revealed by cross-correlated NMR relaxation measurements. J Am Chem Soc 127:828–829
Wijmenga SS, van Buuren BNM (1998) The use of NMR methods for conformational studies of nucleic acids. Prog Nucl Magn Reson Spectrosc 32:287–387
Ying J, Grishaev A, Bax A (2006) Carbon-13 chemical shift anisotropy in DNA bases from field dependence of solution NMR relaxation rates. Magn Reson Chem 44:302–310
Zwahlen C, Vincent SJ (2002) Determination of (1)H homonuclear scalar couplings in unlabeled carbohydrates. J Am Chem Soc 124:7235–7239
Acknowledgements
The paper is dedicated to Christian Griesinger. The work has been supported by the state of Hesse (Center for Biomolecular Magnetic Resonance) and the DFG (Sonderforschungsbereich: RNA-Ligand-Interactions). We wish to thank Oliver Ohlenschläger, Jens Wöhnert and Matthias Görlach for providing us with the 30mer RNA sample.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rinnenthal, J., Richter, C., Ferner, J. et al. Quantitative Γ-HCNCH: determination of the glycosidic torsion angle χ in RNA oligonucleotides from the analysis of CH dipolar cross-correlated relaxation by solution NMR spectroscopy. J Biomol NMR 39, 17–29 (2007). https://doi.org/10.1007/s10858-007-9167-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-007-9167-5