Abstract
Background
Open cast lignite mines, sand pits and military training areas represent human-made, secondary habitats for specialized xerothermophilous and psammophilous species. Rare species, including the earwig Labidura riparia, are found in high population densities in such sites. However, it is unknown from which sources colonisation took place and how genetic variation compares to that of ancient populations on natural sites.
Methods
Using nine microsatellite markers, we analysed genetic variation and population structure of L. riparia in 21 populations in NE Germany both from secondary habitats such as lignite-mining sites, military training areas and a potassium mining heap, and rare primary habitats, such as coastal and inland dunes.
Results
Genetic variation was higher in populations from post-mining sites and former military training areas than in populations from coastal or inland dune sites. Overall population differentiation was substantial (FST = 0.08; FʹST = 0.253), with stronger differentiation among primary (FST = 0.196; FʹST = 0.473) than among secondary habitats (FST = 0.043; FʹST = 0.147). Differentiation followed a pattern of isolation by distance. Bayesian structure analysis revealed three gene pools representing primary habitats on a coastal dune and two different inland dunes. All populations from secondary habitats were mixtures of the two inland dune gene pools, suggesting multiple colonization of post-mining areas from different source populations and hybridisation among source populations.
Discussion
Populations of L. riparia from primary habitats deserve special conservation, because they harbour differentiated gene pools. The majority of the L. riparia populations, however, thrive in secondary habitats, highlighting their role for conservation.
Implications for insect conservation
A dual strategy should be followed of conserving both remaining natural habitat harbouring particular intraspecific gene pools and secondary habitat inhabited by large admixed and genetically highly variable populations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biodiversity is threatened globally and locally by human activities in various ways especially habitat destruction, fragmentation, pollution or climate change (Cardoso et al. 2020). Consequently, nature conservation needs to answer this challenge in multiple targeted ways (Samways et al. 2020). Apart from conserving existing natural and semi-natural habitats, secondary habitats, that also resulted from human action, may play a significant role for practical conservation (e.g. Wiegleb et al. 2013; Torma et al. 2018; Reeves and Daniels 2020).
In Central European glacial lowlands, sand is the most frequent geological substrate. However, nutrient-poor, open and mobile natural sand habitats are rare and confined to marine coasts, river banks and a few unfixed inland dunes. These habitats are naturally free of perennial vegetation due to continuous physical disturbance by water or wind action. Open sand habitats possess a typical fauna, adapted to the special abiotic conditions. Due to the rarity of the habitat type, specialist species of sandy sites find a very limited number of isolated habitats often threatened by anthropogenic habitat destruction.
Lignite open cast mining in Central and East Germany resulted in dumping nutrient-poor sandy substrates over large areas (Hildmann and Wünsche 1996; Wiegleb et al. 2000). Additionally, in military training areas large areas of open sandy soils were created due to vegetation and topsoil destruction by trucks and tanks. In the region of Lower Lusatia open landscapes with a total area of 1000 km2 were generated by anthropogenic influence (Wiegleb and Felinks 2001; Anders et al. 2004). Thus, in Central Europe former mining areas represent rare cases of allowing the study of primary succession at larger scales, as the dumped substrates are initially devoid of higher organisms or diaspores (Prach 1987; Bingemer et al. 2020). In former military training areas secondary succession starting from non-sterile substrates can be observed (Prach and Pysek 2001; Brunk 2007).
Large post-mining landscapes and other disturbed sites offer new habitats for plant and animal species and are rapidly colonised (Meijer 1989; Mrzljak and Wiegleb 2000; Brändle et al. 2003; Walker and del Moral 2003; Brunk et al. 2004; Heneberg et al. 2013). The colonisation of arthropods on such sites and patterns of succession have been intensively studied (e.g. Neumann 1971; Dunger 1991; Brunk 2007). Due to their mere size, the lack of ongoing anthropogenic impact small-scale heterogeneity of abiotic conditions, post-mining landscapes can harbour a rich fauna and flora and can be of high value for conservation (Brändle et al. 2000; Tischew 2004; Dolezalova et al. 2012).
Colonization of previously unoccupied habitats critically depends on the number of colonizing individuals (Brändle et al. 2003). It may involve only few individuals resulting in founder effects, leading to demographic and genetic bottlenecks and reducing genetic variation in founder populations. The genetic diversity of populations is furthermore influenced by the population sizes in patches, the number of patches, and the level of gene flow between patches (Whitlock and Barton 1997). The viability of newly founded populations may be affected negatively by reduced genetic variation (Amos and Balmford 2001; Hansson and Westerberg 2002), especially when environmental conditions change (Reed and Frankham 2003). Therefore, genetic variation and population structure allow both insights into the processes of colonisation and the conservation value of the genetic resources relative to ancient populations on natural sites.
The earwig L. riparia (Pallas, 1773) (Dermaptera: Labiduridae) is an insect species confined to open sandy habitats. In Central Europe, it was previously restricted to a few locations of coastal and inland sand dunes as well as river banks (Zacher 1917; Weidner 1941; Harz 1957). Local populations seem to undergo frequent extinctions (Weidner 1941). However, during the twentieth century the species successfully colonised a large number of secondary sites, like sand pits (Heneberg et al. 2016) and lignite mines characterised by open sand. In Central Europe, L. riparia currently maintains the majority of its population in mining sites (Matzke and Klaus 1996) which thus are important areas for the conservation of this endangered species (Wallaschek 2004). Three principal scenarios may be proposed for the colonisation of anthropogenic sites by L. riparia, given the presence of multiple spatially isolated and putatively genetically differentiated natural populations. First, colonisation of secondary sites could have happened from a single source population resulting in high genetic similarity and either reduced or equal levels of genetic variation in secondary habitats, depending on the strength of bottlenecks. Second, colonisation could have happened from multiple genetically differentiated source populations resulting in a mixture and higher levels of genetic variation than in either source region. Third, since the species is adapted to ephemeral sandy habitat, the newly established sites may have been continuously colonised from a meta-population that existed prior to mining activities, resulting in low levels of genetic differentiation among both old and new populations.
In this study we analysed genetic variation and population structure of L. riparia from primary and secondary habitats in East Germany. We addressed the following questions: (1) Does genetic diversity of earwig populations differ between natural habitats and secondary sites? (2) Are the populations genetically structured, and is genetic differentiation related to geographic distance? (3) Can a single or multiple source regions be identified for colonisation of secondary habitats? (4) What are the consequences for conservation?
Materials and methods
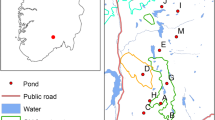
Study species
The sand earwig L. riparia (Pallas, 1773) (Dermaptera: Labiduridae) has a cosmopolitan distribution. It inhabits sandy but damp sites with no or sparse vegetation (Beier 1959; Matzke 2001). In Central and Eastern Europe, the species occurs naturally along the seacoasts, along rivers and on inland dunes. Rivers with open sandy banks were postulated as main former colonization routes in Central Europe. The species was rare and rated ‘highly endangered’ in Germany (Binot et al. 1998). Its historical distribution was limited to a few isolated locations (Fig. 1). Since the middle of the twentieth century, L. riparia was repeatedly found in secondary habitats like open cast mines, dumps or sand and gravel pits in Central and Eastern Germany (Jordan 1957; Donath 1988; Matzke and Klaus 1996; Oelerich 2000; Wallaschek 2004) (Fig. 1). Nowadays, the species also occurs on former military training areas with sandy substrates (Matzke and Klaus 1996). Labidura riparia displays large variation in hind wing length (Weidner 1941). Although flying individuals are only rarely observed, it is assumed that winged individuals are able to disperse (Kleinow 1971; Matzke 1995). Flight ability has also been assumed leading to the rather rapid colonization of new habitats (Matzke and Klaus 1996; Paill 2007). Wing length was measured in all individuals collected in this study and ranged from 1.17 to 2.93 mm in females and 1.20 to 3.55 mm in males. Although it is unclear which wing length allows flying, the large observed length range may indicate that at least some of the individuals are able to fly.
Map of extinct (filled triangle—pre 1941, open circle—1941–1989), recent (filled circle—after 1989) and sampled locations of Labidura riparia in Northeastern Germany. Sampling sites are numbered according to Table 1. Note that site 3 is an inland dune located only 20 km from Baltic coast. Occurrence data according to Zacher (1917), Weidner (1941), Müller-Motzfeld et al. (1990), Köhler and Renker (2001), Klatt (2003), Wallaschek (2004), Matzke (2011) and unpublished recent observations of A. Altenkamp, C. Blumenstein, W. Funk, M. Güth, K. Handke, H. Illig, T. Kappauf
Sampling and genotyping
We sampled 21 sites, 5 in primary habitats on coastal and inland dunes, representing the few known natural populations in NE Germany (Table 1, Fig. 1). Note that site 3, although close to the coast, is an inland dune located 20 km from the Baltic coast without maritime influence (Jeschke 2003). Additionally, 16 sites were sampled in secondary, man-made, habitats such as lignite mines, military training sites and mine waste heaps. While the natural dunes originated in early postglacial times (around 10,000 BC), the age of the secondary habitats was between 2 and 59 years after dumping for the lignite mines and > 60 years for the military training sites. Earwigs were collected in 2004–2006 using one to four live soil traps per location baited with dry dog food. The traps were exposed overnight. Earwigs were sampled in the morning and stored in liquid nitrogen. Sample size ranged from 18 to 38 individuals per site (mean = 25.7).
In total, 539 L. riparia individuals were captured and genotyped at nine microsatellite loci as described previously (Gueth and Durka 2006) with the following modifications. Microsatellite DNA amplifications were carried out in a volume of 10 µL using a multiplex protocol (Qiagen, Hilden, Germany): ca. 20 ng DNA, 2 µL Qiagen multiplex PCR master mix, 0.2 µM of each primer (1 µL) and 2 µL H2O. Two multiplex reactions were performed: Lari05 (label: FAM), Lari10 (JOE), Lari14 (FAM), Lari17 (JOE) in the first reaction and Lari18 (TAMRA), Lari37b (FAM), Lari39b (FAM), Lari51 (FAM) and Lari77 (JOE) in a second reaction. In each locus, some individuals (between 0.2% and 17.4%, average 3.5%, Table 2) did not amplify any fragment, indicating the presence of homozygous null-alleles. Based on the raw data, 15 of the 189 locus × site combinations showed significant departure from Hardy–Weinberg-equilibrium, which also may be due to the presence of null-alleles. Because undetected null-alleles would lead to biased allele frequency estimates, we used the program MicroChecker (Van Oosterhout et al. 2006) to adjust allele frequencies and genotypes assuming Hardy–Weinberg equilibrium MicroChecker suggests for each locus and population a list of genotypes in which null-alleles (“0”) are added to homozygous genotypes according to the estimated null-allele frequency. We subsequently for each locus renamed the suggested null-allele and all homozygous null-alleles as additional allele and thus included them in the analyses. While this adjusted dataset is optimized for allele frequency, the manually modified genotype identities do not allow to calculate inbreeding coefficients. Raw data and Null-allele corrected data are publicly available at https://www.pangaea.de.
Genetic data analysis
Genetic variability at population level was quantified as mean number of alleles across loci (A) and expected heterozygosity (He, gene diversity), which were determined using MSA 3.0 (Dieringer and Schlötterer 2003). Because sample sizes differed among populations, we additionally calculated allelic richness (Ar), a measure of allelic diversity that corrects for sample size, using the program Fstat 2.9.3 (Goudet 1995). Genetic variation among populations was assessed with F-statistics using (Weir and Cockerham 1984) estimators (θ) with the program Fstat. Because microsatellites are highly polymorphic, measures of population differentiation may be biased downward. We therefore used the method of Hedrick (2005) to standardize estimates of population differentiation: \(F^{\prime}_{ST} = {{F_{{{\text{ST}}}} } \mathord{\left/ {\vphantom {{F_{{{\text{ST}}}} } {F_{{{\text{STmax}}}} }}} \right. \kern-\nulldelimiterspace} {F_{{{\text{STmax}}}} }}\). For estimation of FSTmax data were recoded so that each population had a unique set of alleles, keeping the original allele frequencies with the program RecodeData (Meirmans 2006). Arlequin 3.01 (Excoffier et al. 2005) was used to perform analyses of molecular variance (AMOVA) to assess the hierarchical genetic structure of alternative predefined population groupings, i.e. Northern German vs. Central German, primary vs. secondary habitat and old vs. young mining sites. We tested whether genetic differentiation among populations from primary sites differed from that among secondary sites with a permutation procedure in Fstat using 500 permutations. A Mantel test was performed to assess the relationship between genetic differentiation and geographic distance using Arlequin. In order to visualise the relationships among populations we applied principal component analysis (PCA) with PCAGen 1.2 (Goudet 1999).
Furthermore, we applied a model-based Bayesian clustering algorithm to identify gene pools using STRUCTURE 2.3.3 (Pritchard et al. 2000). We used the admixture model with a burnin period of 50,000 and 100,000 iterations and no prior information about population membership. The most probable number of clusters was identified by running the algorithm 20 times each with values of K from 1 to 11 and calculating the posterior probability L(K) and ΔK according to Evanno et al. (2005). Measures of genetic variation were compared among groups of populations using a Tukey HSD-test in R (R Core Team 2018).
Results
Genetic variation within populations
In a total of 539 L. riparia individuals we detected 102 different alleles at 9 microsatellite loci. The number of alleles per locus ranged between 8 (Lari77, Lari51, Lari39b) and 16 in Lari10. Gene diversity ranged between 0.481 (Lari14) and 0.741 in Lari39b (Table 2). All 539 specimens showed different multilocus genotypes. The populations were highly diverse with mean number of alleles ranging from 3.9 to 7.2, and mean A = 5.7, allelic richness ranging from 3.2 to 4.8, with mean Ar = 4.2 and expected heterozygosity ranging from 0.569 to 0.748 with mean He = 0.688.
When genetic variation was compared among habitats, coastal and inland dune sites were less variable than post-mining sites and military training areas with respect to gene diversity and allelic richness (Table 1). Thus, earwig populations from human-made habitat were not genetically impoverished relative to natural habitats but in contrast showed higher levels of diversity. Comparing populations from young (< 17 years) to old (30–59 years) post-mining landscapes did not reveal significant differences in gene diversity, number of alleles or allelic richness (p > 0.3).
Genetic population structure
Populations were significantly structured as revealed by an overall FST value of 0.08 (SD 0.008, p < 0.01). However, as maximal possible differentiation was low due to high allelic diversity (FSTmax = 0.316), the standardised measure of population differentiation indicated substantial overall differentiation, FʹST = 0.253. When this analysis was confined to the primary habitats, the respective values were FST = 0.169 and FʹST = 0.473, indicating pronounced genetic differentiation among natural sites. When confined to secondary anthropogenic sites, the respective values were FST = 0.043 and FʹST = 0.147, which differed significantly from primary habitats (p = 0.002), indicating that populations in human-made sites are much less differentiated than natural populations. Analysis of molecular variance (AMOVA) similarly indicated significant differentiation among populations with 7.9% of molecular variance residing among populations (p = 0.001). Further hierarchical AMOVAs were carried out for different groupings of sites (Table 3). The most pronounced differentiation was found for Northern German (sites 1–3) vs. Central German (4–21) sites. A lower, but significant differentiation was found for primary habitats (coastal plus inland dunes) vs. secondary habitats. No differentiation was found for post-mining sites vs. military areas and young vs. old post-mining sites. In all these AMOVAs, differentiation among populations within groups was stronger than differentiation among groups.
Out of 210 pairwise FST values, 202 were significant (p < 0.05). The population structure was consistent with a model of isolation by distance as revealed by a significant correlation between genetic differentiation and geographic distance (Fig. 2, Mantel-test p < 0.001), indicating equilibrium between gene flow and genetic drift and limited long-distance dispersal.
Individual-based analyses
Principal component analysis showed that populations from natural habitats on coastal and inland dunes were separated from secondary sites in post-mining and military training sites which formed one cluster (Fig. 3). Interestingly, population 3 (Altwarp) from an inland dune located near the Baltic coast was clearly separated from the other coastal populations (1, 2, Hiddensee). While these coastal populations where clearly distinct and separated from all inland populations along the first PCA axis, those from the two inland dunes where both closer to the cluster of secondary sites which took an intermediate position between these two inland dunes.
PCA of microsatellite allele frequencies of Labidura riparia populations from natural, primary sites along the Baltic coast (filled triangles), natural inland dunes (filled squares) and from secondary, anthropogenic sites (open circles). Numbers refer to sites (Table 1)
The PCA results were fully corroborated by Bayesian cluster analysis which revealed three gene pools (Fig. 4). The three gene pools correspond to three groups of natural populations, which were characterized by population level membership coefficients ≥ 95% (Fig. 5). A fourth group was made up by secondary populations which showed admixture of gene pools. The first cluster included individuals from the coastal populations (site 1/2) and the second and third cluster those from the two inland dune systems 3 (Altwarp) and 4/5 (Klein Schmölen), respectively. Populations from secondary habitats mostly showed mixture and individual admixture of the gene pools of the two inland dune gene pools. The contribution of these two inland dune clusters in the populations on secondary habitat varied between 20.2–87.4% (site 3) and 9.9–78.3% (site 4/5) while the coastal gene pool contributed to negligible extent (1.5–19.2%, mean 4.9%).
Mean posterior probability (Ln P(D)) and ΔK as function of number of clusters, K, in STRUCTURE analyses according to Evanno et al. (2005) resulting in K = 3 as most probable number of gene pools for 539 L. riparia individuals from 21 sites
Discussion
We showed that genetic variation in L. riparia populations was higher in secondary sites than in the natural primary sites studied. The geographically isolated primary populations on coastal and inland dunes were clearly differentiated from one another, while the populations on secondary sites showed close relationships to those on the inland dunes. The Bayesian cluster analysis showed that secondary sites were of mixed origin suggesting colonization of post-mining areas from multiple inland dunes and further internal spread.
Colonisation history
During the ice ages, L. riparia presumably withdrew into refuges in East, Southeast and Southwest Europe (Weidner 1941). After the retreat of the glaciers, two colonisation pathways to the north were possible. The melting water rivers with southeast-north-western direction are generally proposed routes on which earwigs and other biota migrated starting from south-east Europe to northern Germany colonising suitable habitats on inland dunes and finally on coastal regions (Harz 1957; Adis and Junk 2002). Although our data do not provide clear evidence on postglacial migration routes, the genetic differentiation among coastal and inland does not preclude the existence of several independent colonization pathways.
Open sandy habitats must have been much more common in early postglacial times and were important habitat for many arthropod groups. During this period L. riparia presumably formed large populations harbouring high levels of genetic variation. Later on, the species presumably was repressed into isolated sites on coastal and inland dunes. The strong genetic differentiation between the gene pools representing the natural populations in Germany (FST = 0.196, FʹST = 0.473) suggests strongly restricted gene flow among these populations. In these isolated sites genetic drift may have reduced genetic diversity which is lowest in the populations from the Baltic coast (Table 1). Therefore, prior to increased mining activities starting in the second half of the nineteenth century, L. riparia was rare and putatively affected by genetic drift in spatially isolated or ephemeral habitats. First recordings of single L riparia individuals in Central German museum collections date back to this period (Halle: 1869; Thuringia: 1873, Köhler and Renker 2001; Wallaschek 2004). The species was found in secondary habitats in the early twentieth century on military training sites which existed for 200 years, but with lower impact on habitat structures (Ramme 1911). Other early authors listed single sites where Labidura had been observed, and simultaneously, where it was no longer found at that time (Zacher 1917; Weidner 1941). This indicates that suitable habitats for the species must have been rare and unstable prior to large scale anthropogenic disturbance. Only after open cast lignite mining created large areas of open sandy soils starting during the first half of the twentieth century, L. riparia colonised the newly developed habitats, thereby drastically increasing its population. Although two natural populations from inland dunes were identified as putative source of the secondary populations, we cannot rule out the existence of additional source populations that may also have contributed to the colonisation of secondary habitats.
Genetic differentiation and mobility
Gene flow among insect populations and thus the degree of genetic differentiation is determined, among others, by the dispersal power of insect species. The overall genetic differentiation of L. riparia (FST = 0.08, FʹST = 0.253) was similar to that of other arthropod species with either high mobility and/or large local population sizes, like e.g. common ground beetles Carabus auronitens: FST = 0.075 (Drees 2003) and Pogonus chalceus, FST = 0.041 (Desender et al. 1998)), butterflies like Maculinea nausithous: FST = 0.068, FʹST = 0.158 (Anton et al. 2007) or grasshoppers (Lange et al. 2010). Stronger genetic population structure was only found in flightless habitat specialists confined to spatially isolated habitat patches, e.g. in forest-dwelling Carabus-species [e.g. Carabus variolosus, FST = 0.465, (Matern et al. 2009) C. auronitens, FST = 0.49 (Desender et al. 2002)]. Genetic differentiation of L. riparia was only slightly higher than found in the common European earwig, Forficula auriculata [FST = 0.055, (Brown 2006)]. Both earwig species are winged but have been observed flying very rarely. However, despite their apparently poor ability to disperse by flight, F. auriculata is very easily transported by human activity due to its synanthropic crevice-seeking behaviour (Brown 2006). Although such behaviour is uncommon in L. riparia, the role of anthropogenic transport cannot be ruled out completely, e.g. with transport of sand or mining and military equipment. The comparison with other taxa and its cosmopolitan distribution suggests that although being a habitat specialist, L. riparia is relatively well dispersed thereby maintaining gene flow. However, FST values based on microsatellite data depend on gene diversity and may strongly underestimate genetic differentiation (Hedrick 2005). In fact, standardized genetic differentiation in L. riparia was FʹST = 0.253 overall and FʹST = 0.147 for the anthropogenic sites, which indicate founder effects and suggest that colonisation of new sites may be accomplished by only a small number of animals.
Implications for conservation
The natural habitats on coastal and inland dunes harbouring L. riparia populations were small and spatially isolated. There, populations probably are much smaller than in large continuous secondary sites. The natural populations are highly endangered by environmental stochasticity, demographic changes and genetic drift. This is underlined by the extinction of populations on inland river banks during the last century (Weidner 1941, Fig. 1). On the other hand, the Baltic coast population on Hiddensee Island is known to have persisted at the very same location since the 1950s (Günther 1971). It is an open question whether the reduced genetic variation in this population is responsible for the fact that it does neither increase nor colonize new sites along the Baltic coastline. The populations on primary coastal and inland dunes formed three differentiated gene pools. These populations deserve special conservation in future as they may represent locally adapted gene pools. It would be interesting to investigate whether the low genetic variation in primary sites compared to admixed populations on secondary sites has consequences for the fitness of these populations.
From the perspective of nature conservation, successional post-mining areas, sand and gravel pits as well as military training areas are refuge habitats for many threatened taxa including xerothermo- and psammophilous species (e.g. Brändle et al. 2000; Warren and Buttner 2008; Heneberg et al. 2013, 2016). Labidura riparia has profited from the expansion of such sites in the past and has developed large and genetically diverse populations. Genetic variation was higher in secondary sites due to admixture of genotypes from several source populations. These newly established populations in turn can act as source populations or stepping stones during colonisation of new habitats. Early successional habitats will inevitably undergo changes and will be overgrown by dense vegetation on the long run (Wiegleb and Felinks 2001; Felinks and Wiegand 2008). Thus, populations on secondary sites are potentially endangered, unless physical disturbance continues, creating new open habitats and allowing for the development of a metapopulation structure with repeated extinction and recolonization events.
References
Adis J, Junk WJ (2002) Terrestrial invertebrates inhabiting lowland river floodplains of Central Amazonia and Central Europe: a review. Freshwat Biol 47:711–731
Amos W, Balmford A (2001) When does conservation genetics matter? Heredity 87:257–265
Anders K, Mrzljak J, Wallschläger D, Wiegleb G (2004) Handbuch Offenlandmanagement am Beispiel ehemaliger und in Nutzung befindlicher Truppenübungsplätze. Springer, Heidelberg
Anton C, Zeisset I, Musche M, Durka W, Boomsma JJ, Settele J (2007) Population structure of a large blue butterfly and its specialist parasitoid in a fragmented landscape. Mol Ecol 16:3828–3838
Beier M (1959) Ordnung: Dermaptera. In: Weber H (ed) Bronn’s Klassen und Ordnungen des Tierreichs, vol V. Akademische Verlagsgesellschaft, Leipzig, pp 455–586
Bingemer J, Pfeiffer M, Hohberg K (2020) First 12 years of tardigrade succession in the young soils of a quickly evolving ecosystem. Zool J Linn Soc 188:887–899
Binot M, Bless R, Boye P, Gruttke H, Pretscher P (1998) Rote Liste gefährdeter Tiere Deutschlands. Bundesamt für Naturschutz, Bonn
Brändle M, Durka W, Altmoos M (2000) Biodiversity of surface dwelling beetles in open-cast lignite mines in Central Germany. Biodivers Conserv 9:1297–1311
Brändle M, Durka W, Krug H, Brandl R (2003) The assembly of a local flora and fauna: plants and birds in non-reclaimed mining sites. Ecography 26:652–660
Brown GS (2006) Sperm competition and male forceps dimorphism in the European earwig Forficula auricularia (Dermaptera: Forficulina)
Brunk I (2007) Diversität und Sukzession von Laufkäferzönosen in gestörten Landschaften Südbrandenburgs. BTU Cottbus
Brunk I, Mrzljak J, Wiegleb G (2004) Recultivation of post-mining areas with Quercus rubra—the succession of ground beetle communities. Mitt Dtsch Ges Allg Angew Ent 14:323–326
Cardoso P, Barton PS, Birkhofer K, Chichorro F, Deacon C, Fartmann T, Fukushima CS, Gaigher R, Habel JC, Hallmann CA, Hill MJ, Hochkirch A, Kwak ML, Mammola S, Noriega JA, Orfinger AB, Pedraza F, Pryke JS, Roque FO, Settele J, Simaika JP, Stork NE, Suhling F, Vorster C, Samways MJ (2020) Scientists’ warning to humanity on insect extinctions. Biol Conserv. https://doi.org/10.1016/j.biocon.2020.108426
Desender K, Backeljau T, Delahaye K, De Meester L (1998) Age and size of European saltmarshes and the population genetic consequences for ground beetles. Oecologia 114:503–513
Desender K, Verdyck P, Gaublomme E, Dhuyvetter H, Rasplus JY (2002) Extreme genetic differentiation and isolation by nondistance in Carabus auronitens in relation to forest historical ecology in Flanders (Belgium). In: Bauer T, Den Boer PJ, Lövei G, Szyszko J (eds) How to protect and what we know about Carabid Beetles. Warsaw Agricultural Press, Warsaw, pp 227–235
Dieringer D, Schlötterer C (2003) Microsatellite Analyser (MSA): a platform independent analysis tool for large microsatellite data sets. Mol Ecol Notes 3:167–169
Dolezalova J, Vojar J, Smolova D, Solsky M, Kopecky O (2012) Technical reclamation and spontaneous succession produce different water habitats: a case study from Czech post-mining sites. Ecol Eng 43:5–12
Donath H (1988) Der Sandohrwurm (Labidura riparia, PALL.) in der nordwestlichen Niederlausitz Beiträge zur Insektenfauna der nordwestlichen Niederlausitz. Biol Studienreihe Luckau 17:31–35
Drees C (2003) Ausbreitung flugunfähiger Arthropoden über Habitatkorridore—Untersuchungen an einer Metapopulation von Carabus auronitens (Col., Carabidae) im Münsterland Münster
Dunger W (1991) Zur Primärsukzession humiphager Tiergruppen auf Bergbauflächen. Zoologi Jahrb für System 118–3(4):423–447
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evolut Bioinform Online 1:47–50
Felinks B, Wiegand T (2008) Exploring spatiotemporal patterns in early stages of primary succession on former lignite mining sites. J Veg Sci 19:267–276
Goudet J (1995) FSTAT v–1.2: a computer program to calculate F-statistics. J Hered 86:485–486
Goudet J (1999) PCA-GEN for windows V 1.2 (Institute of Ecology, University of Lausanne, Lausanne).http://www.unilch/popgen/softwares/pcagen.html
Gueth M, Durka W (2006) Identification of 10 microsatellite loci in the earwig Labidura riparia (Dermaptera, Labiduridae). Mol Ecol Notes 6:877–879
Günther KK (1971) Die Geradflügler Mecklenburgs (Orthopteroidea und Blattoidea). Faun. Abh. Mus. Tierk. Dresden 3:159–179
Hansson B, Westerberg L (2002) On the correlation between heterozygosity and fitness in natural populations. Mol Ecol 11:2467–2474
Harz K (1957) Die Geradflügler Mitteleuropas. Fischer, Jena
Hedrick PW (2005) A standardized genetic differentiation measure. Evolution 59:1633–1638
Heneberg P, Bogusch P, Rehounek J (2013) Sandpits provide critical refuge for bees and wasps (Hymenoptera: Apocrita). J Insect Conserv 17:473–490
Heneberg P, Hesoun P, Skuhrovec J (2016) Succession of arthropods on xerothermophilous habitats formed by sand quarrying: epigeic beetles (Coleoptera) and orthopteroids (Orthoptera, Dermaptera and Blattodea). Ecol Eng 95:340–356
Hildmann E, Wünsche M (1996) Lignite mining and its after-effects on the Central German landscape. Water Air Soil Pollut 91:79–87
Jeschke L (2003) Altwarper Binnendünen, Neuwarper See und Riether Werder 186. In: Mecklenburg-Vorpommern U (ed) Die Naturschutzgebiete in Mecklenburg-Vorpommern. Demmler-Verlag, Schwerin, p 206
Jordan KH (1957) Die Ohrwürmer der Niederlausitz. Nachr der Oberlausitzer Insektenfreunde 5:2–3
Klatt R (2003) Ein selten werdender Kosmopolit: Der Sandohrwurm Labidura riparia. Naturschutz-Förderverein „Döberitzer Heide“ eV. Beitr zum Nat, zur Landsch und zur Gesch 13:27–31
Kleinow W (1971) Morphometrische Untersuchungen an Flugapparaten flugfähiger Dermapteren. Zool Anz 187:175–184
Köhler G, Renker C (2001) Beitrag zu einer Fauna der Ohrwürmer (Insecta: Dermaptera) Thüringens. Thüring Faunist Abh 8:63–81
Lange R, Durka W, Holzhauer SIJ, Wolters V, Diekötter T (2010) Differential threshold effects of habitat fragmentation on gene flow in two widespread species of bush crickets. Mol Ecol 19:4936–4948
Matern A, Desender K, Drees C, Gaublomme E, Paill W, Assmann T (2009) Genetic diversity and population structure of the endangered insect species Carabus variolosus in its western distribution range: implications for conservation. Conserv Genet 10:391–405
Matzke D (1995) Bemerkenswerte Beobachtungen und Funde des Sandohrwurms Labidura riparia in Tagebauen und Sandgruben bei Leipzig. Entomol Nach und Ber 39:91–92
Matzke D (2001) Verzeichnis der Ohrwürmer (Dermaptera) Deutschlands. Entomofauna Ger 5:53–59
Matzke D (2011) Fauna der Ohrwürmer (Dermaptera) und Schaben (Blattoptera) Sachsens. In: Klausnitzer B, Reinhardt R (eds) Beiträge zur Insektenfauna Sachsens Vol 9. Mitteilungen Sächsischer Entomologen Supplement pp 9–81
Matzke D, Klaus D (1996) Zum Vorkommen des Sandohrwurms (Labidura riparia PALLAS) auf Abgrabungsflächen Nordwest-Sachsens und angrenzender Gebiete (Insecta, Dermaptera, Labiduridae). Mauritiana 16:13–19
Meijer J (1989) Sixteen years of fauna invasion and succession in the Lauwerzeepolder. In: Majer JD (ed) Animals in primary succession. Cambridge University Press, Cambirdge, pp 339–369
Meirmans PG (2006) Using the AMOVA framework to estimate a standardized genetic differentiation measure. Evolution 60:2399–2402
Mrzljak J, Wiegleb G (2000) Spider colonization of former brown coal mining areas—time or structure dependent? Landsc Urb Plan 51:131–146
Müller-Motzfeld G, Niemann A, Mathyl E (1990) Im Rahmen der Küsteninselkartierung erfasste Käfer (Coleoptera) und Ohrenkriecher (Dermaptera). Nat und Umwelt, Beitr aus dem Bez Rostock 15:17–56
Neumann U (1971) Die Sukzession der Bodenfauna (Carabidae—Coleoptera, Diplopoda u. Isopoda) in den forstlich rekultivierten Gebieten des Rheinischen Braunkohlereviers. Pedobiol 11:193–226
Oelerich HM (2000) Zur Geradflüglerfauna der Braunkohlen-Bergbaufolgelandschaften Sachsen-Anhalts (Dermaptera). Herz NF 33:117–154
Paill WG (2007) Neue Funde des Sandohrwurms Labidura riparia (PALLAS, 1773) in Wien und aus dem Südburgenland (Dermaptera). Beitr zur Entomofaunist 8:149–152
Prach K (1987) Succession of vegetation on dumps from strip coal mining, N.W. Bohemia. Czechoslo Folia Geobot Phytotaxon 22:339–354
Prach K, Pysek P (2001) Using spontaneous succession for restoration of human-disturbed habitats: experience from Central Europe. Ecol Eng 17:55–62
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
R Core Team (2018) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Ramme W (1911) Ein Beitrag zur Kenntnis der Orthopterenfauna der Mark Brandenburg. Berl Entomol Ztg 56:1–10
Reed DH, Frankham R (2003) Correlation between fitness and genetic diversity. Conserv Biol 17:230–237
Reeves LE, Daniels JC (2020) Conservation value of secondary forest habitats to endemic frugivorous butterflies at Mount Kanlaon, Negros Occidental, Philippines. J Insect Conserv 24:913–926
Samways MJ, Barton PS, Birkhofer K, Chichorro F, Deacon C, Fartmann T, Fukushima CS, Gaigher R, Habel JC, Hallmann CA, Hill MJ, Hochkirch A, Kaila L, Kwak ML, Maes D, Mammola S, Noriega JA, Orfinger AB, Pedraza F, Pryke JS, Roque FO, Settele J, Simaika JP, Stork NE, Suhling F, Vorster C, Cardoso P (2020) Solutions for humanity on how to conserve insects. Biol Conserv 242:108427
Tischew S (ed) (2004) Renaturierung nach dem Braunkohleabbau. Teubner, Stuttgart, Leipzig, Wiesbaden
Torma A, Bozsó M, Gallé R (2018) Secondary habitats are important in biodiversity conservation: a case study on orthopterans along ditch banks. Anim Biodivers Conserv 41:97–108
Van Oosterhout C, Weetman D, Hutchinson WF (2006) Estimation and adjustment of microsatellite null alleles in nonequilibrium populations. Mol Ecol Notes 6:255–256
Walker LR, del Moral R (2003) Primary succession and ecosystem rehabilitation. Cambridge University Press, Cambridge
Wallaschek M (2004) Rote Liste der Ohrwürmer (Dermaptera) des Landes Sachsen-Anhalts. In:Wallaschek M, Langer T, Richter K (eds) Die Geradflügler des Landes Sachsen-Anhalt Heuschrecken Ohrwürmer Fangschrecken und Schaben Berichte des Landesamtes für Umweltschutz Sachsen-Anhalt 5 pp 220–222
Warren SD, Buttner R (2008) Active military training areas as refugia for disturbance-dependent endangered insects. J Insect Conserv 12:671–676
Weidner H (1941) Vorkommen und Lebensweise des Sandohrwurms, Labidura riparia Pall. Zool Anz 133:185–202
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Whitlock MC, Barton NH (1997) The effective size of a subdivided population. Genetics 146:427–441
Wiegleb G, Felinks B (2001) Predictability of early stages of primary succession in post-mining landscapes of Lower Lusatia, Germany. Appl Veg Sci 4:5–18
Wiegleb G, Bröring U, Mrzljak J, Schulz F (2000) Naturschutz in Bergbaufolgelandschaften—Landschaftsanalyse und Leitbildentwicklung. Physica, Heidelberg
Wiegleb G, Broring U, Choi G, Dahms HU, Kanongdate K, Byeon CW, Ler LG (2013) Ecological restoration as precaution and not as restitutional compensation. Biodivers Conserv 22:1931–1948
Zacher F (1917) Die Geradflügler Deutschlands und ihre Verbreitung. G. Fischer, Jena
Acknowledgements
We are highly indebted to Danilo Matzke and Günter Köhler for sharing their experience and samples, to Filip Niemann, Barbara Seidl-Lampa, Gisela Koch, Ingo Brunk, Karl Heyde, Ingolf Stodian and Heiko Sparmberger who helped sampling Labidura, to Cornelia Baeßler and Jörn Vorwald for help with the distribution map and to Thorsten Assmann and Fabian Haas for comments to earlier versions of the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was supported by the Federal Ministry of Education and Research (Grant No. BMBF-FKz 01LC0018A) within the framework of SUBICON (Successional Change and Biodiversity Conservation) and by the European Fond for Regional Development.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gueth, M., Wiegleb, G. & Durka, W. Colonisation of secondary habitats in mining sites by Labidura riparia (Dermaptera: Labiduridae) from multiple natural source populations. J Insect Conserv 25, 349–359 (2021). https://doi.org/10.1007/s10841-021-00305-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10841-021-00305-y