Abstract
An activity cliff (AC) is formed by a pair of structurally similar compounds with a large difference in potency. Accordingly, ACs reveal structure–activity relationship (SAR) discontinuity and provide SAR information for compound optimization. Herein, we have investigated the question if ACs could be predicted from image data. Therefore, pairs of structural analogs were extracted from different compound activity classes that formed or did not form ACs. From these compound pairs, consistently formatted images were generated. Image sets were used to train and test convolutional neural network (CNN) models to systematically distinguish between ACs and non-ACs. The CNN models were found to predict ACs with overall high accuracy, as assessed using alternative performance measures, hence establishing proof-of-principle. Moreover, gradient weights from convolutional layers were mapped to test compounds and identified characteristic structural features that contributed to successful predictions. Weight-based feature visualization revealed the ability of CNN models to learn chemistry from images at a high level of resolution and aided in the interpretation of model decisions with intrinsic black box character.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In recent years, convolutional neural networks (CNNs) have gained increasing attention in chemical informatics and pharmaceutical research. For example, two-dimensional (2D) images of molecular graphs [1,2,3,4,5] and three-dimensional (3D) images of activity landscapes [6] have been used for deriving CNN models and extracting specific features from image data. For example, the Inception-ResNet v2 architecture was used to train CNN models on images from a large data set comprising 1.7 million compounds and predict physicochemical properties such as logP [1]. In addition, quantitative property predictions on the basis of compound images were reported using Chemception [2] and ChemNet [3]. Furthermore, the Toxic Colors approach [4] added atom labels, colored dots, and partial charge maps to image representations for compound toxicity predictions while Kekulescope [5] only used Kekulé structures as input for compound potency and cell line toxicity predictions. Taken together, these investigations have indicated the potential of various CNN architectures to extract specified molecular features from 2D image representations and use these features for property predictions. Different from molecular structure-based approaches, 3D images of activity landscape variants were used for feature extraction and classification of landscape models according to structure–activity relationship (SAR) characteristics of the corresponding compound data sets [6].
While CNNs have thus far mostly been trained on 2D compound images, to our knowledge, they have not been used to process images of pairs of closely related compounds and predict differences in properties at the level of pairs. Activity cliffs (ACs) represent a prominent paradigm for compound pair-encoded property differences [7]. ACs are defined as pairs or groups of similar compounds or structural analogs with large differences in activity (potency) [7, 8]. Accordingly, ACs embody the pinnacle of SAR discontinuity, i.e., small chemical modifications leading to large potency alterations, and are a major source of SAR information [8]. An elegant formalism for the systematic identification of pairs of structural analogs is the matched molecular pair (MMP) concept and its algorithmic implementation [9]. An MMP is defined as a pair of compounds that share a common core structure and are only distinguished by a chemical modification at a single site (termed a chemical transformation) [9]. As such, MMPs are well suited for representing ACs, which has led to the introduction of MMP-cliffs [10]. An MMP-cliff is defined as an MMP formed by two compounds that are active against the same target and have a statistically significant difference in potency [10].
As a consistent molecular representation, MMP-cliffs have been used for predicting ACs at different levels. First, MMP-cliffs have been systematically distinguished from MMPs with only small or no potency differences using support vector machine classification on the basis of fingerprint representations and specialized compound pair-based kernel functions [11]. Subsequently, MMP-cliffs have also been successfully predicted in a methodologically simpler manner applying the condensed graph of reaction formalism [12]. In addition, potency differences encoded by MMPs have been quantitatively predicted using support vector regression [13]. To aid in the interpretation of machine learning models, fingerprint features determining correct AC predictions have been mapped back to the original compounds to delineate critically important substructures distinguishing MMP-cliffs from other MMPs [11].
Herein, we have attempted to predict MMP-cliffs from image data using CNNs. In addition to assessing classification performance, we have made use of recent developments in convolutional layer visualization [14,15,16,17] to identify and display key features contributing to correct AC predictions. Our proof-of-concept investigation further extends the current spectrum of molecular image-based modeling in chemical informatics.
Material and methods
Compound activity classes
From ChEMBL (version 26) [18], three compound activity classes with available high-confidence activity data were extracted. Compounds were tested against single human targets in direct interaction assays at highest assay confidence (ChEMBL confidence score 9). As potency measurements, assay-independent equilibrium constants (pKi values) were required. Multiple measurements for the same compound were averaged, provided all values fell within the same order of magnitude; otherwise, the compound was disregarded. Table 1 reports the targets and composition of these activity classes.
Matched molecular pairs and activity cliffs
For activity classes, all possible MMPs were generated by systematically fragmenting individual exocyclic single bonds and sampling core structures and substituents in index tables [9]. For substituents, size restrictions were applied to limit MMP formation to typically observed structural analogs [10]. Accordingly, a substituent was permitted to contain at most 13 non-hydrogen atoms and the core had to be at least twice as large as the substituent. Additionally, for MMP compounds, the maximum difference in non-hydrogen atoms between the substituents was set to eight, yielding transformation size-restricted MMPs [10].
An MMP qualified as an MMP-cliff if the two structural analogs had an at least 100-fold difference in potency (ΔpKi ≥ 2.0) [10]. To avoid potency difference-dependent boundary effects in AC prediction, compounds forming a non-AC MMP were restricted to an at most tenfold difference in potency. Furthermore, to balance structural heterogeneity of large activity classes originating from different sources, MMPs were only retained if their compounds and core structures were found in multiple MMPs. Table 1 reports MMP and MMP-cliff statistics for the activity classes.
Molecular image representations
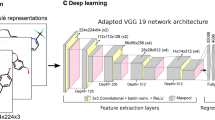
Each MMP core and the associated substituents were treated as separate molecular objects using the RDKit application programming interface (API) [19]. For each unique core and substituent, high-resolution portable network graphics (PNG) compound images with 500 × 500 pixels were generated using the RDkit Chem.Draw package (version 2020.03.5) [19]. In images, substituent attachment sites were replaced with an asterisk symbol. To represent an MMP core and the two substituents defining the transformation in combined form, core and substituent images were resized to 300 × 300 pixels and then horizontally concatenated in a single image of dimensions 300 × 900 × 3 (height × width × color-channels). Figure 1 illustrates MMP image generation. The pixel values of all image matrices were converted into 32-bit floating point format and normalized. Images were processed using openCV (version 4.4.0) [20,21,22].
Convolutional neural network architecture
Figure 2 shows the CNN architecture designed for image analysis, consisting of convolutional, pooling, dropout, and dense layers. Two convolutional layers with kernel size of 32 and respective filter sizes of 3 × 3 and 5 × 5 were used to extract key features from MMP images. The convolutional layers were followed by a pooling, dropout, and dense layer. Max-pooling was used as pooling layer to compute the maximum value in each patch of each convolved feature map. A dropout layer was added to avoid overfitting. After ‘flattening’ the weights, the softmax function was applied to normalize learned weights and yield a probability distribution. CNN layers were implemented using TensorFlow (version 2.2.0) [23] and Keras (version 2.2.4) [24].
Performance measures
CNN models were trained to systematically distinguish between MMP-cliffs and corresponding non-AC MMPs. The classification performance of CNN models was evaluated using receiver-operator characteristic (ROC) curves and the area under the ROC curve (AUC). In addition, model performance was assessed with four performance measures including overall accuracy (A), balanced accuracy (BA), weighted mean F1 score [25], and Mathews correlation coefficient (MCC) [26], defined as:
TP, TN, FP, and FN denote true positives, true negatives, false positives, and false negatives respectively.
Convolutional layer feature visualization
Spatial information from the convolutional layers of trained models was extracted using the Grad-Cam algorithm [17]. Channel-based mean values of the resulting convolutional feature map activation weights were mapped to the original image for feature visualization.
Results and discussion
Convolutional neural network models
CNN models were derived to distinguish between MMP-cliffs and non-AC MMPs on the basis of molecular images generated for three distinct activity classes including thrombin inhibitors (target/activity class ID 204), tyrosine kinase Abl inhibitors (class 1862), and mu opioid receptor ligands (class 233). As shown in Fig. 1, MMP images for CNNs combined the shared core structure with the pair of substituents representing the chemical transformation. Images of the three structures constituting an MMP were concatenated horizontally to obtain a single image. In contrast to displaying two compounds forming an MMP side-by-side, this image format contained no redundant substructure (duplicated core).
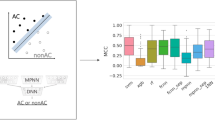
CNN models were separately trained in 10 independent trials on a set of 4050–10,178 images, dependent on the activity class. Training images were obtained by randomly selecting half of the MMP-cliffs per class (228–561; Table 1) and half of the non-AC MMPs (1797–4856). The resulting models were then tested on the remaining half of the MMP-cliff and non-AC MMP images. ROC curves for the best performing individual classification models are shown in Fig. 3. These CNN models yielded accurate AC predictions, with ROC-AUC values of 0.97 (204), 0.93 (233), and 0.92 (1862). In addition, Table 2 reports the mean prediction accuracy of the CNN models for each activity class on the basis of alternative performance measures. Although training and test sets were imbalanced, i.e., they containing many more non-AC MMPs than MMP-cliffs, the predictions were generally stable (i.e., yielding very low standard deviations) and consistently successful on the basis of all performance measures. Overall, CNN classification accuracy was highest for thrombin inhibitors (class 204), with mean AUC of 0.97 (AUC = 0.97), F1 = 0.85, MCC = 0.83, A = 0.97, and BA = 0.90, followed by class 1862 and 233. Although AUC and A values were also high for class 233 (0.92 and 0.96, respectively), predictions for this class yielded lowest F1 = 0.36, MCC = 0.39, and BA = 0.63 values, indicating that the majority class (non-AC MMPs) was predicted here more accurately than the minority class (MMP-cliffs). In this case, only < 5% of all MMPs represented MMP-cliffs. Thus, these results were expected. The use of balanced training sets would likely further increase prediction accuracy, which is meaningful from a machine learning perspective. However, for AC predictions, balancing MMP-cliff and non-AC MMP training sets would represent an unrealistic scenario because ACs are generally rare among qualifying compound pairs [8]. Regardless, even in the presence of class label imbalance, image-based classification of MMP-cliffs vs. non-AC MMPs was overall surprisingly accurate, more so than we anticipated.
Image feature visualization
Convolutional features naturally retain spatial information, which is lost in fully-connected layers. Therefore, the Grad-Cam algorithm was applied to visualize convolutional layer activation weights [17]. Figures 4, 5 and 6 show examples of original images onto which channel-based mean values of activation weights of the corresponding convolutional feature map were superimposed. All MMPs shown in Figs. 4 and 5 were correctly predicted while Fig. 6 also shows a false positive prediction. Visualization of convolutional layers revealed that most of the key image features were captured by the first convolutional layer. However, in a number of instances, the second convolutional layer was also capable of extracting and emphasizing key features, as shown in Fig. 7. Accordingly, addition of the second convolution layer typically further improved classification accuracy.
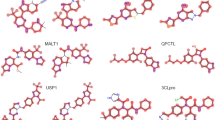
Mapping of activation weights. For four exemplary MMPs from class 204, mean gradient weights of the first convolutional layer are displayed on the respective structures and color-coded according to the given continuous color spectrum. Classification probabilities for each class (AC, non-AC) are given (%) and the correct class label of each MMP is colored in yellow. Shown are a a non-AC MMP with phenyl and cyclohexyl substituents, b and c MMP-cliffs with similar core structures and substituents, and d a non-AC MMP with different aliphatic ring substituents
Mapping of activation weights. For three similar MMPs from class 204, mean gradient weights of the first convolutional layer are displayed. The representation is according to Fig. 4. a and b show MMP-cliffs and c shows a non-AC MMP. Highly weighted secondary or tertiary amines are encircled
Mapping of activation weights. For three MMPs from class 204, mean gradient weights of the first convolutional layer are displayed. The representation is according to Fig. 4. a and b show a correctly predicted MMP-cliff and non-AC MMP, respectively. Highly weighted primary or secondary amines are encircled. c shows a false positive MMP-cliff prediction. Highly weighted primary and secondary amines are shared by the substituents. The distinguishing single and double bonds were detected with medium weights and are encircled
Mapping of first and second layer activation weights. a and b show the MMPs from Figs. 5c and 6c, respectively, with mean gradient weights of the first and second convolutional layer. Here, weights from the second convolution layer reinforced structural features detected by first layer weights and also identified additional features
Learning structural features from compound images
The convolutional layer weights of the best performing CNN model (class 204) for MMP images from the test set were systematically extracted and visualized. A compelling observation was that weights from the CNN models detected specific structural features in MMP images. For example, convolutional layers were capable of recognizing primary, secondary, and tertiary amines as well as various ring structures. Moreover, the model was able to differentiate between substituents with different structures. In Fig. 4, the model distinguished between ring and aliphatic substituents, which is clearly evident by comparing mapped convolutional layer weights. Different weight distributions led to accurate predictions of MMP-cliffs and non-AC MMPs with very high probabilities of at least 94%. Furthermore, the model learned to differentiate between alternative cyclic structures, hence accounting for molecular topology.
In Fig. 5, the CNN model assigned high weights to secondary and tertiary amines in rings of substituents of correctly predicted MMP-cliffs and non-AC MMPs. Notably, the presence of different amines was a characteristic feature of all MMPs originating from class 204. However, by comparing the MMP-cliff and non-AC MMP in Fig. 5b and c, respectively, it becomes clear that detecting a tertiary amine alone was not sufficient to distinguish between the MMP-cliff and non-AC MMP because this feature was shared by both. In this case, the core structures of these MMPs were distinct and different core features were detected and assigned high weights, hence illustrating core contributions to accurate predictions. In the MMPs shown in Fig. 6a and b, primary and secondary amines were also detected as distinguishing features in aliphatic substructures. Furthermore, Fig. 6c reports a false positive MMP-cliff prediction. On the basis of the MMP alone, this prediction error cannot be rationalized. To these ends, weights in similar MMPs with different class labels must be compared, as illustrated in Fig. 5. Nonetheless, this example was interesting because the replacement of a single bond with a double bond, i.e., a change in bond order representing a minute chemical modification at the level of images, was detected with medium weights as a distinguishing substituent feature.
Taken together, these convolutional layer weight-based visualizations demonstrated the capacity of the CNN model to detect signature features of compounds from a given activity class (such as the presence of various amines) as well as specific chemical features that distinguished cores and/or substituents of MMPs, including different ring structures, individual functional groups, or bond orders. Mapping weights from different convolutional layers often further emphasized such features or identified additional ones, as illustrated in Fig. 7. The correct detection of specific features distinguishing MMPs with different class labels provided a rationale for the overall accuracy of the AC predictions. Differences between substituents detected by the CNN model can be analyzed at the level of individual MMP images, while understanding differently weighted core features requires comparisons of multiple MMPs. Visualization of key features in MMP cores and substituents aids in the interpretation of CNN model decisions that typically have black box character, hence improving model accessibility.
Conclusion
In the work, we have attempted the prediction of MMP-cliffs, which are an intuitive AC representation, on the basis of MMP image data using CNN models. To our knowledge, these are the first molecular image-based property predictions at the level of compound pairs. In our proof-of-concept investigation, encouraging accuracy was achieved in systematically distinguishing between MMP-cliffs and non-AC MMPs. While ACs were successfully predicted before using other machine learning approaches, we have been particularly interested in the question whether CNNs are capable of extracting chemical features and small feature differences from images of pairs of structural analogs that correctly distinguish between SAR continuity (embodied by non-ACs) and discontinuity (ACs). Mapping of convolutional layer weights to test compounds and visualizing corresponding structural features put the analysis on a level beyond statistical assessment of prediction accuracy. Visualization revealed the ability of CNN models to detect specific chemical features including distinct substructures and individual functional groups that distinguished structural analogs or MMPs with different properties. Thus, the models were capable to learn chemistry from MMP images, which resulted in successful AC predictions.
References
Szegedy C, Ioffe S, Vanhoucke V, Alemi A (2016) Inception-v4, Inception-ResNet and the impact of residual connections on learning. arXiv:1602.07261
Goh GB, Siegel C, Vishnu A, Hodas NO, Baker N (2017) Chemception: a deep neural network with minimal chemistry knowledge matches the performance of expert-developed QSAR/QSPR models. arXiv:1706.06689
Goh GB, Vishnu A, Siegel C, Hodas N (2018) Using rule-based labels for weak supervised learning: a ChemNet for transferable chemical property prediction. In: Proceedings of the ACM SIGKDD international conference on knowledge discovery and data mining
Fernandez M, Ban F, Woo G, Hsing M, Yamazaki T, LeBlanc E, Rennie PS, Welch WJ, Cherkasov A (2018) Toxic Colors: the use of deep learning for predicting toxicity of compounds merely from their graphic images. J Chem Inf Model 58:1533–1543
Cortés-Ciriano I, Bender A (2019) KekuleScope: prediction of cancer cell line sensitivity and compound potency using convolutional neural networks trained on compound images. J Cheminform 11:e41
Iqbal J, Vogt M, Bajorath J (2020) Activity landscape image analysis using convolutional neural networks. J Cheminform 12:e34
Maggiora GM (2006) On outliers and activity cliffs—why QSAR often disappoints. J Chem Inf Model 46:1535–1535
Stumpfe D, Bajorath J (2012) Exploring activity cliffs in medicinal chemistry. J Med Chem 55:2932–2942
Hussain J, Rea C (2010) Computationally efficient algorithm to identify matched molecular pairs (MMPs) in large data sets. J Chem Inf Model 50:339–348
Hu X, Hu Y, Vogt M, Stumpfe D, Bajorath J (2012) MMP-Cliffs: systematic identification of activity cliffs on the basis of matched molecular pairs. J Chem Inf Model 52:1138–1145
Heikamp K, Hu X, Yan A, Bajorath J (2012) Prediction of activity cliffs using support vector machines. J Chem Inf Model 52:2354–2365
Horvath D, Marcou G, Varnek A, Kayastha S, de la Vega de León A, Bajorath J (2016) Prediction of activity cliffs using condensed graphs of reaction representations, descriptor recombination, support vector machine classification, and support vector regression. J Chem Inf Model 56:1631–1640
de la Vega de León A, Bajorath J (2014) Prediction of compound potency changes in matched molecular pairs using support vector regression. J Chem Inf Model 54:2654–2663
Griffin G, Perona P (2008) Learning and using taxonomies for fast visual categorization. In: 2008 IEEE conference on computer vision and pattern recognition. pp 1–8
Mahendran A, Vedaldi A (2016) Visualizing deep convolutional neural networks using natural pre-images. Int J Comput Vis 120:233–255
Nguyen A, Yosinski J, Clune J (2016) Multifaceted feature visualization: uncovering the different types of features learned by each neuron in deep neural networks. arXiv:1602.03616
Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D (2017) Grad-CAM: visual explanations from deep networks via gradient-based localization. In: 2017 ieee international conference on computer vision (ICCV). pp 618–626
Gaulton A, Hersey A, Nowotka ML, Patricia Bento A, Chambers J, Mendez D, Mutowo P, Atkinson F, Bellis LJ, Cibrian-Uhalte E, Davies M, Dedman N, Karlsson A, Magarinos MP, Overington JP, Papadatos G, Smit I, Leach AR (2017) The ChEMBL database in 2017. Nucleic Acids Res 45:D945–D954
Landrum G RDKit: open-source cheminformatics. https://www.rdkit.org. Accessed 19 Jan 2021
Culjak I, Abram D, Pribanic T, Dzapo H, Cifrek M (2012) A brief introduction to OpenCV. In: MIPRO 2012—35th international convention on information and communication technology, electronics and microelectronics—proceedings. pp 1725–1730
OpenCv (2014) OpenCV library. https://opencv.org. Accessed 19 Jan 2021
Bradski G (2000) The OpenCV library. Dr Dobb’s J Softw Tools 25:120–125
Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M, Kudlur M, Levenberg J, Monga R, Moore S, Murray DG, Steiner B, Tucker P, Vasudevan V, Warden P, Wicke M, Yu Y, Zheng X. (2016) TensorFlow: a system for large-scale machine learning. In: 12th USENIX Symposium on operating systems design and implementation (OSDI 16), Savannah, GA
Chollet F (2015) Keras. https://github.com/keras-team/keras. Accessed 19 Jan 2021
Chinchor N (1992) MUC-4 evaluation metrics. In: Proceedings of the 4th conference on message understanding. Association for Computational Linguistics, USA. pp 22–29
Matthews BW (1975) Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim Biophys Acta 405:442–451
Acknowledgements
J.I. is supported by a PhD fellowship from the German Academic Exchange Service (DAAD) in collaboration with the Higher Education Commission (HEC) of Pakistan.
Funding
Open Access funding enabled and organized by Projekt DEAL..
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Iqbal, J., Vogt, M. & Bajorath, J. Prediction of activity cliffs on the basis of images using convolutional neural networks. J Comput Aided Mol Des 35, 1157–1164 (2021). https://doi.org/10.1007/s10822-021-00380-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-021-00380-y