Abstract
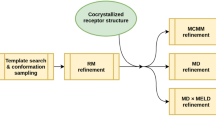
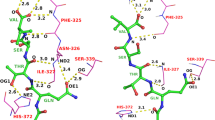
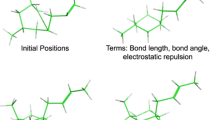
The D3R Grand Challenge 4 provided a brilliant opportunity to test macrocyclic docking protocols on a diverse high-quality experimental data. We participated in both pose and affinity prediction exercises. Overall, we aimed to use an automated structure-based docking pipeline built around a set of tools developed in our team. This exercise again demonstrated a crucial importance of the correct local ligand geometry for the overall success of docking. Starting from the second part of the pose prediction stage, we developed a stable pipeline for sampling macrocycle conformers. This resulted in the subangstrom average precision of our pose predictions. In the affinity prediction exercise we obtained average results. However, we could improve these when using docking poses submitted by the best predictors. Our docking tools including the Convex-PL scoring function are available at https://team.inria.fr/nano-d/software/.






Similar content being viewed by others
References
Jusot M, Stratmann D, Vaisset M, Chomilier J, Cortes J (2018) Exhaustive exploration of the conformational landscape of small cyclic peptides using a robotics approach. J Chem Inf Model 58(11):2355–2368
Sindhikara D, Spronk SA, Day T, Borrelli K, Cheney DL, Posy SL (2017) Improving accuracy, diversity, and speed with prime macrocycle conformational sampling. J Chem Inf Model 57(8):1881–1894
Smith RD, Damm-Ganamet KL, Dunbar JB Jr, Ahmed A, Chinnaswamy K, Delproposto JE, Kubish GM, Tinberg CE, Khare SD, Dou J et al (2015) Csar benchmark exercise 2013: evaluation of results from a combined computational protein design, docking, and scoring/ranking challenge. J Chem Inf Model 56(6):1022–1031
Carlson HA, Smith RD, Damm-Ganamet KL, Stuckey JA, Ahmed A, Convery MA, Somers DO, Kranz M, Elkins PA, Cui G, Peishoff CE, Lambert MH, Dunbar JB Jr (2016) Csar 2014: a benchmark exercise using unpublished data from pharma. J Chem Inf Model 56(6):1063–1077
Gathiaka S, Liu S, Chiu M, Yang H, Stuckey JA, Kang YN, Delproposto J, Kubish G, Dunbar JB, Carlson HA et al (2016) D3R grand challenge 2015: evaluation of protein–ligand pose and affinity predictions. J. Comput. Aided Mol. Des. 30(9):651–668
Gaieb Z, Liu S, Gathiaka S, Chiu M, Yang H, Shao C, Feher VA, Walters WP, Kuhn B, Rudolph MG et al (2018) D3R grand challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies. J Comput Aided Mol Des 32(1):1–20
Gaieb Z, Parks CD, Chiu M, Yang H, Shao C, Walters WP, Lambert MH, Nevins N, Bembenek SD, Ameriks MK et al (2019) D3R grand challenge 3: blind prediction of protein-ligand poses and affinity rankings. J Comput Aided Mol Des 33(1):1–18
Rose PW, Prlić A, Altunkaya A, Bi C, Bradley AR, Christie CH, Di Costanzo L, Duarte JM, Dutta S, Feng Z et al (2017) The rcsb protein data bank: integrative view of protein, gene and 3d structural information. Nucleic Acids Res 45(D1):D271–D281
Ignatov M, Liu C, Alekseenko A, Sun Z, Padhorny D, Kotelnikov S, Kazennov A, Grebenkin I, Kholodov Y, Kolosvari I et al (2019) Monte carlo on the manifold and md refinement for binding pose prediction of protein-ligand complexes: 2017 D3R grand challenge. J Comput Aided Mol Des 33(1):119–127
Kumar A, Zhang KY (2019) Shape similarity guided pose prediction: lessons from D3R grand challenge 3. J Comput Aided Mol Des 33(1):47–59
Koukos PI, Xue LC, Bonvin AM (2019) Protein-ligand pose and affinity prediction: lessons from D3R grand challenge 3. J Comput Aided Mol Des 33(1):83–91
Nguyen DD, Cang Z, Wu K, Wang M, Cao Y, Wei G-W (2019) Mathematical deep learning for pose and binding affinity prediction and ranking in D3R grand challenges. J Comput Aided Mol Des 33(1):71–82
Lam PC-H, Abagyan R, Totrov M (2019) Hybrid receptor structure/ligand-based docking and activity prediction in icm: development and evaluation in D3R grand challenge 3. J Comput Aided Mol Des 33(1):35–46
Chaput L, Selwa E, Elisee E, Iorga BI (2019) Blinded evaluation of cathepsin s inhibitors from the D3RGC3 dataset using molecular docking and free energy calculations. J Comput Aided Mol Des 33(1):93–103
Sunseri J, King JE, Francoeur PG, Koes DR (2019) Convolutional neural network scoring and minimization in the D3R 2017 community challenge. J Comput Aided Mol Des 33(1):19–34
He X, Man VH, Ji B, Xie X-Q, Wang J (2019) Calculate protein-ligand binding affinities with the extended linear interaction energy method: application on the cathepsin s set in the D3R grand challenge 3. J Comput Aided Mol Des 33(1):105–117
Xie B, Minh DD (2019) Alchemical grid dock (algdock) calculations in the D3R grand challenge 3. J Comput Aided Mol Des 33(1):61–69
Vassar R, Kovacs DM, Yan R, Wong PC (2009) The \(\beta\)-secretase enzyme bace in health and alzheimer’s disease: regulation, cell biology, function, and therapeutic potential. J Neurosci 29(41):12787–12794
Prati F, Bottegoni G, Bolognesi ML, Cavalli A (2017) Bace-1 inhibitors: from recent single-target molecules to multitarget compounds for alzheimer’s disease: Miniperspective. J Med Chem 61(3):619–637
Hanessian S, Yang G, Rondeau J-M, Neumann U, Betschart C, Tintelnot-Blomley M (2006) Structure-based design and synthesis of macroheterocyclic peptidomimetic inhibitors of the aspartic protease \(\beta\)-site amyloid precursor protein cleaving enzyme (bace). J Med Chem 49(15):4544–4567
Jordan JB, Whittington DA, Bartberger MD, Sickmier EA, Chen K, Cheng Y, Judd T (2016) Fragment-linking approach using 19f nmr spectroscopy to obtain highly potent and selective inhibitors of \(\beta\)-secretase. J Med Chem 59(8):3732–3749
Butini S, Brogi S, Novellino E, Campiani G, Ghosh AK, Brindisi M, Gemma S (2013) The structural evolution of \(\beta\)-secretase inhibitors: a focus on the development of small-molecule inhibitors. Curr Top Med Chem 13(15):1787–1807
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461
Kadukova M, Grudinin S (2017) Convex-pl: a novel knowledge-based potential for protein-ligand interactions deduced from structural databases using convex optimization. J Comput Aided Mol Des 31(10):943–958
Kadukova M, Grudinin S (2016) Knodle: a support vector machines-based automatic perception of organic molecules from 3d coordinates. J Chem Inf Model 56:1410–9
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) Autodock4 and autodocktools4: automated docking with selective receptor flexibility. J Comput Chem 30(16):2785–2791
Liu Z, Su M, Han L, Liu J, Yang Q, Li Y, Wang R (2017) Forging the basis for developing protein-ligand interaction scoring functions. Acc Chem Res 50(2):302–309
Artemova S, Grudinin S, Redon S (2011) A comparison of neighbor search algorithms for large rigid molecules. J Comput Chem 32(13):2865–2877
Klenin KV, Tristram F, Strunk T, Wenzel W (2011) Derivatives of molecular surface area and volume: simple and exact analytical formulas. J Comput Chem 32(12):2647–2653
Klenin K, Tristram F, Strunk T, Wenzel W (2012) Achieving numerical stability in analytical computation of the molecular surface and volume. From Computational Biophysics to Systems Biology (CBSB11)–Celebrating Harold Scheraga’s 90th Birthday 8:75
Grudinin S, Popov P, Neveu E, Cheremovskiy G (2015) Predicting binding poses and affinities in the csar 2013–2014 docking exercises using the knowledge-based convex-pl potential. J Chem Inf Model 56(6):1053–1062
Grudinin S, Kadukova M, Eisenbarth A, Marillet S, Cazals F (2016) Predicting binding poses and affinities for protein-ligand complexes in the 2015 D3R grand challenge using a physical model with a statistical parameter estimation. J Comput Aided Mol Des 30(9):791–804
Kadukova M, Grudinin S (2018) Docking of small molecules to farnesoid x receptors using autodock vina with the convex-pl potential: lessons learned from D3R grand challenge 2. J Comput Aided Mol Des 32(1):151–162
G Landrum Rdkit: Open-source cheminformatics http://www.rdkit.org
Riniker S, Landrum GA (2015) Better informed distance geometry: using what we know to improve conformation generation. J Chem Inf Model 55(12):2562–2574
Tosco P, Stiefl N, Landrum G (2014) Bringing the MMFF force field to the rdkit: implementation and validation. J Cheminform 6(1):37
Schrödinger, LLC (2011) The PyMOL molecular graphics system, version 1.3
Acknowledgements
The authors would like to thank Ivan Gushchin from MIPT Moscow for providing his expertise in crystallography. This work was partially supported by the Ministry of Education and Science of the Russian Federation (Grant No. 6.3157.2017).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kadukova, M., Chupin, V. & Grudinin, S. Docking rigid macrocycles using Convex-PL, AutoDock Vina, and RDKit in the D3R Grand Challenge 4. J Comput Aided Mol Des 34, 191–200 (2020). https://doi.org/10.1007/s10822-019-00263-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-019-00263-3