Abstract
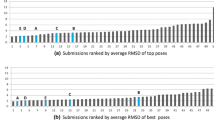
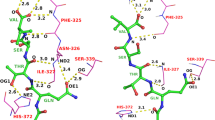
The 2015 D3R Grand Challenge provided an opportunity to test our new model for the binding free energy of small molecules, as well as to assess our protocol to predict binding poses for protein-ligand complexes. Our pose predictions were ranked 3–9 for the HSP90 dataset, depending on the assessment metric. For the MAP4K dataset the ranks are very dispersed and equal to 2–35, depending on the assessment metric, which does not provide any insight into the accuracy of the method. The main success of our pose prediction protocol was the re-scoring stage using the recently developed Convex-PL potential. We make a thorough analysis of our docking predictions made with AutoDock Vina and discuss the effect of the choice of rigid receptor templates, the number of flexible residues in the binding pocket, the binding pocket size, and the benefits of re-scoring. However, the main challenge was to predict experimentally determined binding affinities for two blind test sets. Our affinity prediction model consisted of two terms, a pairwise-additive enthalpy, and a non pairwise-additive entropy. We trained the free parameters of the model with a regularized regression using affinity and structural data from the PDBBind database. Our model performed very well on the training set, however, failed on the two test sets. We explain the drawback and pitfalls of our model, in particular in terms of relative coverage of the test set by the training set and missed dynamical properties from crystal structures, and discuss different routes to improve it.






Similar content being viewed by others
References
Smith RD, Dunbar JJB, Ung PM, Esposito EX, Yang CY, Wang S, Carlson HA (2011) J Chem Inf Model 51:2115
Damm-Ganamet KL, Smith RD, Dunbar JB Jr, Stuckey JA, Carlson HA (2013) J Chem Inf Model 53(8):1853
Grudinin S, Popov P, Neveu E, Cheremovskiy G (2015) J Chem Inf Model. doi:10.1021/acs.jcim.5b00339
Crawford TD, Ndubaku CO, Chen H, Boggs JW, Bravo BJ, Delatorre K, Giannetti AM, Gould SE, Harris SF, Magnuson SR, McNamara E, Murray LJ, Nonomiya J, Sambrone A, Schmidt S, Smyczek T, Stanley M, Vitorino P, Wang L, West K, Wu P, Ye W (2014) J Med Chem 57(8):3484. doi:10.1021/jm500155b
Homeyer N, Gohlke H (2013) J Comput Chem 34(11):965
Wang L, Wu Y, Deng Y, Kim B, Pierce L, Krilov G, Lupyan D, Robinson S, Dahlgren MK, Greenwood J et al (2015) J Am Chem Soc 137(7):2695
Wang L, Berne B, Friesner RA (2012) PNAS 109(6):1937
Lensink MF, Velankar S, Kryshtafovych A, Huang SY, Schneidman-Duhovny D, Sali A, Segura J, Fernandez-Fuentes N, Viswanath S, Elber R, Grudinin S, Popov P, Neveu E, Lee H, Baek M, Park S, Heo L, Rie G, Lee C Seok, Qin S, Zhou HX, Ritchie DW, Maigret B, Devignes MD, Ghoorah A, Torchala M, Chaleil RAG, Bates PA, Ben-Zeev E, Eisenstein M, Negi SS, Weng Z, Vreven T, Pierce BG, Borrman TM, Yu J, Ochsenbein F, Guerois R, Vangone A, Rodrigues JPGLM, van Zundert G, Nellen M, Xue L, Karaca E, Melquiond ASJ, Visscher K, Kastritis PL, Bonvin AMJJ, Xu X, Qiu L, Yan C, Li J, Ma Z, Cheng J, Zou X, Shen Y, Peterson LX, Kim HR, Roy A, Han X, Esquivel-Rodriguez J, Kihara D, Yu X, Bruce NJ, Fuller JC, Wade RC, Anishchenko I, Kundrotas PJ, Vakser IA, Imai K, Yamada K, Oda T, Nakamura T, Tomii K, Pallara C, Romero-Durana M, Jiménez-García B, Moal IH, JFérnandez-Recio IH, Joung JY, Kim JY, Joo K, Lee J, Kozakov D, Vajda S, Mottarella S, Hall DR, Beglov D, Mamonov A, Xia B, Bohnuud T, Del Carpio CA, Ichiishi E, Marze N, Kuroda D, Roy Burman SS, Gray JJ, Chermak E, Cavallo L, Oliva R, Tovchigrechko A, Wodak SJ (2016) Proteins. doi:10.1002/prot.25007
Popov P, Grudinin S (2015) J Chem Inf Model 55(10):2242. doi:10.1021/acs.jcim.5b00372
Marillet S, Boudinot P, Cazals F (2015) Proteins: Struct Funct Bioinform 1(84): 9 (2015). doi:10.1002/prot.24946. https://hal.inria.fr/hal-01159641
Kastritis P, Moal I, Hwang H, Weng Z, Bates P, Bonvin A, Janin J (2011) Protein Sci 20:482
Huang SY, Zou X (2008) Proteins: Struct Funct Bioinform 72(2):557http://onlinelibrary.wiley.com/doi/10.1002/prot.21949/full
Chuang GY, Kozakov D, Brenke R, Comeau SR, Vajda S (2008) Biophys J 95(9):4217
Maiorov VN, Grippen GM (1992) J Mol Biol 227(3):876
Qiu J, Elber R (2005) Proteins: Struct Funct Bioinform 61(1):44
Rajgaria R, McAllister S, Floudas C (2006) Proteins: Struct Funct Bioinform 65(3):726
Tobi D, Bahar I (2006) Proteins: Struct Funct Bioinform 62(4):970
Ravikant D, Elber R (2010) Proteins: Struct Funct Bioinform 78(2):400
Chae MH, Krull F, Lorenzen S, Knapp EW (2010) Proteins: Struct Funct Bioinform 4(78):1026
Neudert G, Klebe G (2011) Bioinformatics 27(7):1021
Conte L Lo, Chothia C, Janin J (1999) JMB 285(5):2177
Janin J, Bahadur RP, Chakrabarti P (2008) Q Rev Biophysics 41(2):133
Cazals F, Proust F, Bahadur R, Janin J (2006) Protein Sci 15(9):2082. doi:10.1110/ps.062245906
Loriot S, Cazals F (2010) Bioinformatics 26(7):964. doi:10.1093/bioinformatics/btq052. http://hal.inria.fr/hal-00849822
Gerstein M, Richards F (2001) Protein geometry: volumes, areas, and distances. In: Rossmann MG, Arnold E (eds) The international tables for crystallography, vol F, Chap 22. Springer, Berlin, p 531–539
Cazals F, Kanhere H, Loriot S (2011) ACM Trans Math Softw 38(1):1. doi:10.1145/2049662.2049665. http://hal.inria.fr/hal-00849809
Meng G, Arkus N, Brenner M, Manoharan V (2010) Science 327(5965):560
Dunitz J (1995) Chem Biol 2(11):709
Kastritis P, Rodrigues J, Folkers G, Boelens R, Bonvin A (2014) JMB 426:2632
Eisenberg D, Wesson M, Yamashita M (1989) Chem Scr A 29:217
Bouvier B, Grunberg R, Nilgès M, Cazals F (2009) Proteins: Struct Funct Bioinform 76(3):677. doi:10.1002/prot.22381. http://hal.inria.fr/hal-00796032
Wang R, Fang X, Lu Y, Yang CY, Wang S (2005) J Med Chem 48(12):4111. doi:10.1021/jm048957q. http://www.ncbi.nlm.nih.gov/pubmed/15943484
Wang R, Fang X, Lu Y, Wang S (2004) J Med Chem 47(12):2977. doi:10.1021/jm030580l. http://www.ncbi.nlm.nih.gov/pubmed/15163179
Barber D (2012) Bayesian reasoning and machine learning. Cambridge University Press, Cambridge. http://assets.cambridge.org/97805215/18147/cover/9780521518147.jpg
Sonnenburg S, Rätsch G, Henschel S, Widmer C, Behr J, Zien A, Bona Fd, Binder A, Gehl C, Franc V (2010) J Mach Learn Res 11:1799
Kadukova M, Grudinin S (2016) J Chem Inf Model 56(8):1410. doi:10.1021/acs.jcim.5b00512
Trott O, Olson AJ (2010) J Comput Chem 31(2):455
Seeliger D, de Groot BL (2010) J Comput Aided Mol Des 24(5):417. doi:10.1007/s10822-010-9352-6
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) J Cheminform 3:33
Heifets A, Lilien R (2010) J Mol Graph Model 29(1):93. doi:10.1016/j.jmgm.2010.05.005
The PyMOL Molecular Graphics System, Version 1.7 Schrödinger, LLC
Györfi L, Krzyzak A (2002) A distribution-free theory of nonparametric regression. Springer, Berlin
Acknowledgments
The authors thank Dr. Petr Popov from MIPT Moscow for the initial analysis of the HSP90 targets.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Grudinin, S., Kadukova, M., Eisenbarth, A. et al. Predicting binding poses and affinities for protein - ligand complexes in the 2015 D3R Grand Challenge using a physical model with a statistical parameter estimation. J Comput Aided Mol Des 30, 791–804 (2016). https://doi.org/10.1007/s10822-016-9976-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-016-9976-2