Abstract
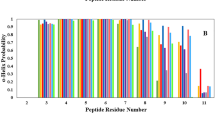
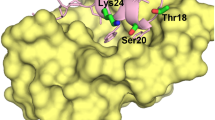
The p53 protein activation protects the organism from propagation of cells with damaged DNA having oncogenic mutations. In normal cells, activity of p53 is controlled by interaction with MDM2. The well understood p53-MDM2 interaction facilitates design of ligands that could potentially disrupt or prevent the complexation owing to its emergence as an important objective for cancer therapy. However, thermodynamic quantification of the p53-peptide induced structural changes of the MDM2-protein remains an area to be explored. This study attempts to understand the conformational free energy and entropy costs due to this complex formation from the histograms of dihedral angles generated from molecular dynamics simulations. Residue-specific quantification illustrates that, hydrophobic residues of the protein contribute maximum to the conformational thermodynamic changes. Thermodynamic quantification of structural changes of the protein unfold the fact that, p53 binding provides a source of inter-element cooperativity among the protein secondary structural elements, where the highest affected structural elements (α2 and α4) found at the binding site of the protein affects faraway structural elements (β1 and Loop1) of the protein. The communication perhaps involves water mediated hydrogen bonded network formation. Further, we infer that in inhibitory F19A mutation of P53, though Phe19 is important in the recognition process, it has less prominent contribution in the stability of the complex. Collectively, this study provides vivid microscopic understanding of the interaction within the protein complex along with exploring mutation sites, which will contribute further to engineer the protein function and binding affinity.









Similar content being viewed by others
References
Vogelstein B, Lane D, Levine AJ (2000) Surfing the p53 network. Nature 408:307–310
El-Deiry WS (1998) Regulation of p53 downstream genes. Semin Cancer Biol 8:345–357
Hermeking H, Eick D (1994) Mediation of c-Myc-induced apoptosis by p53. Science 265:2091–2093
Wu X, Levine AJ (1994) p53 and E2F-1 cooperate to mediate apoptosis. Proc Natl Acad Sci USA 91:3602–3606
Harris CC (1996) p53 tumor suppressor gene: from the basic research laboratory to the clinic–an abridged historical perspective. Carcinogenesis 17:1187–1198
Greenblatt MS, Bennett WP, Hollstein M, Harris CC (1994) Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res 54:4855–4878
Reifenberger G, Liu L, Ichimura K, Schmidt EE, Collins VP (1993) Amplification and overexpression of the MDM2 gene in a subset of human malignant gliomas without p53 mutations. Cancer Res 53:2736–2739
Oliner JD et al (1992) Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 358:80–83
Bueso-Ramos CE, Yang Y, DeLeon E, McCown P, Stass SA, Albitar M (1993) The human MDM-2 oncogene is overexpressed in leukemias. Blood 82:2617–2623
Marchetti A et al (1995) mdm2 gene alterations and mdm2 protein expression in breast carcinomas. J Pathol 175:31–38
Espinoza-Fonseca LM, Trujillo-Ferrara JG (2006) Conformational changes of the p53-binding cleft of MDM2 revealed by molecular dynamics simulations. Biopolymers 83:365–373
Kussie PH et al (1996) Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 274:948–953
Dastidar SG, Raghunathan D, Nicholson J, Hupp TR, Lane DP, Verma CS (2011) Chemical states of the N-terminal ‘lid’ of MDM2 regulate p53 binding: simulations reveal complexities of modulation. Cell Cycle 10:82–89
Dastidar SG, Madhumalar A, Fuentes G, Lane DP, Verma CS (2010) Forces mediating protein–protein interactions: a computational study of p53 ‘approaching’ MDM2. Theor Chem Acc 125:621–635
Chen J, Marechal V, Levine AJ (1993) Mapping of the p53 and mdm-2 interaction domains. Mol Cell Biol 13:4107–4114
Verkhivker GM, Bouzida D, Gehlhaar DK, Rejto PA, Freer ST, Rose PW (2003) Simulating disorder-order transitions in molecular recognition of unstructured proteins: where folding meets binding. Proc Natl Acad Sci U S A 100:5148–5153
Trizac E, Levy Y, Wolynes PG (2010) Capillarity theory for the fly-casting mechanism. Proc Natl Acad Sci USA 107:2746–2750
Lacy ER et al (2004) p27 binds cyclin–CDK complexes through a sequential mechanism involving binding-induced protein folding. Nat Struct Mol Biol 11:358–364
Dastidar SG, Lane DP, Verma CS (2012) Why is F19Ap53 unable to bind MDM2? Simulations suggest crack propagation modulates binding. Cell Cycle 11:2239–2247
Chen J (2009) Intrinsically disordered p53 extreme C-terminus binds to S100B(ββ) through fly-casting. J Am Chem Soc 131:2088–2089
Gsponer J, Madan M, Babu (2009) The rules of disorder or why disorder rules. Prog Biophys Mol Biol 99:94–103
Uhrinova S et al (2005) Structure of free MDM2 N-terminal domain reveals conformational adjustments that accompany p53-binding. J Mol Biol 350:587–598
Dastidar SG, Lane DP, Verma CS (2009) Modulation of p53 binding to MDM2: computational studies reveal important roles of Tyr100. BMC Bioinformatics 10:S6
Zhong H, Carlson HA (2005) Computational studies and peptidomimetic design for the human p53-MDM2 complex. Proteins 58:222–234
Li C et al (2010) Systematic mutational analysis of peptide inhibition of the p53-MDM2/MDMX interactions. J Mol Biol 398:200–213
Buolamwini JK, Addo J, Kamath S, Patil S, Mason D, Ores M (2005) Small molecule antagonists of the MDM2 oncoprotein as anticancer agents. Curr Cancer Drug Targets 5:57–68
Nicholson J, Hupp TR (2010) The molecular dynamics of MDM2. Cell Cycle 9:1878–1881
Portefaix JM et al (2000) Critical residues of epitopes recognized by several anti-p53 monoclonal antibodies correspond to key residues of p53 involved in interactions with the mdm2 protein. J Immunol Methods 244:17–28
Kortemme T, Baker D (2002) A simple physical model for binding energy hot spots in protein–protein complexes. Proc Natl Acad Sci USA 99:14116–14121
Massova I, Kollman PA (1999) Computational alanine scanning to probe protein–protein interactions: a novel approach to evaluate binding free energies. J Am Chem Soc 121:8133–8143
Almerico AM, Tutone M, Pantano L, Lauria A (2012) Molecular dynamics studies on Mdm2 complexes: an analysis of the inhibitor influence. Biochem Biophys Res Commun 424:341–347
Wasylyk C et al (1999) p53 mediated death of cells overexpressing MDM2 by an inhibitor of MDM2 interaction with p53. Oncogene 18:1921–1934
Böttger A et al (1997) Molecular characterization of the hdm2-p53 interaction. J Mol Biol 269:744–756
García-Echeverría C, Chène P, Blommers M. J. J., Furet P (2000) Discovery of potent antagonists of the interaction between human double minute 2 and tumor suppressor p53. J Med Chem 43:3205–3208
Guo Z, Streu K, Krilov G, Mohanty U (2014) Probing the origin of structural stability of single and double stapled p53 peptide analogs bound to MDM2. Chem. Biol Drug Des 83:631–642
Fry DC, Vassilev LT (2005) Targeting protein–protein interactions for cancer therapy. J Mol Med 83:955–963
Arkin M (2005) Protein–protein interactions and cancer: small molecules going in for the kill. Curr Opin Chem Biol 9:317–324
Chène P (2004) Inhibition of the p53-hdm2 interaction with low molecular weight compounds. Cell Cycle 3:458–459
Chène P (2004) Inhibition of the p53-MDM2 interaction: targeting a protein–protein interface. Mol Cancer Res 2:20–28
ElSawy K, Verma CS, Joseph TL, Lane DP, Twarock R, Caves L (2013) On the interaction mechanisms of a p53 peptide and nutlin with the MDM2 and MDMX proteins: a Brownian dynamics study. Cell Cycle 12:394–404
Phan J et al (2010) Structure-based design of high affinity peptides inhibiting the interaction of p53 with MDM2 and MDMX. J Biol Chem 285:2174–2183
Samanta S, Mukherjee S (2017) Microscopic insight into thermodynamics of conformational changes of SAP-SLAM complex in signal transduction cascade. J Chem Phys 146:165103–165111
Karplus M, McCammon JA (2002) Molecular dynamics simulations of biomolecules. Nat Struct Biol 9:646–652
Zerze GH, Uz B, Mittal J (2015) Folding thermodynamics of -hairpins studied by replica-exchange molecular dynamics simulations. Proteins 83:1307–1315
Dror RO, Dirks RM, Grossman JP, Xu H, Shaw DE (2012) Biomolecular simulation: a computational microscope for molecular biology. Annu Rev Biophys 41:429–452
Aldeghi M et al (2016) Accurate calculation of the absolute free energy of binding for drug molecules. Chem Sci 7:207–218
Das A, Gur M, Cheng MH, Jo S, Bahar I, Roux B (2014) Exploring the conformational transitions of biomolecular systems using a simple two-state anisotropic network model. PLoS Comput Biol 10:e1003521
Ovchinnikov V, Cecchini M, Karplus M (2013) A simplified confinement method for calculating absolute free energies and free energy and entropy differences. J Phys Chem B 117:750–762
Esque J, Cecchini M (2015) Accurate calculation of conformational free energy differences in explicit water: the confinement–solvation free energy approach. J Phys Chem B 119:5194–5207
Maragakis P, Spichty AM, Karplus M (2008) A differential fluctuation theorem. J Phys Chem B 112:6168–6174
Das A, Chakrabarti J, Ghosh M (2013) Conformational contribution to thermodynamics of binding in protein–peptide complexes through microscopic simulation. Biophys J 104:1274–1284
McQuarrie DA, Statistical mechanics. (Harper & Row, New York, 1975)
Li D-W, Meng D, Brüschweiler R (2009) Short-range coherence of internal protein dynamics revealed by high-precision in silico study. J Am Chem Soc 131:14610–14611
Trbovic N et al (2009) Protein side-chain dynamics and residual conformational entropy. J Am Chem Soc 131:615–622
Sikdar S, Chakrabarti J, Ghosh M (2014) A microscopic insight from conformational thermodynamics to functional ligand binding in proteins. Mol Biosyst 10:3280–3289
Phillips JC et al (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26:1781–1802
Best RB et al (2012) Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J Chem Theory Comput 8:3257–3273
Ryckaert J-P, Ciccotti G, Berendsen HJ (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23:327–341
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: An N ⋅log(N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092
Allen MP, Tildesley DJ. Computer simulation of liquids. (Clarendon, Oxford, 1987)
Surmiak E et al (2016) A unique Mdm2-binding mode of the 3-pyrrolin-2-one- and 2-furanone-based antagonists of the p53-Mdm2 interaction. ACS Chem Biol 11:3310–3318
Bista M et al (2013) Transient protein states in designing inhibitors of the MDM2-p53 interaction. Structure 21:2143–2151
Acknowledgements
S.S. thanks the National Research Foundation of Singapore for support through BioSym Interdisciplinary Research group at Singapore-MIT Alliance for Research and Technology center.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Samanta, S., Mukherjee, S. Co-operative intra-protein structural response due to protein–protein complexation revealed through thermodynamic quantification: study of MDM2-p53 binding. J Comput Aided Mol Des 31, 891–903 (2017). https://doi.org/10.1007/s10822-017-0057-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-017-0057-y