Abstract
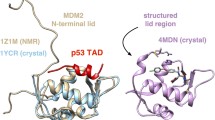
The protein MDM2 forms a complex with the tumor suppressing protein p53 and targets it for proteolysis in order to down-regulate p53 in normal cells. Inhibition of this interaction is of therapeutic importance. Molecular dynamics simulations of the association between p53 and MDM2 have revealed mutual modulation of the two surfaces. Analysis of the simulations of the two species approaching each other in solution shows how long range electrostatics steers these two proteins together. The net electrostatics is controlled largely by a few cationic residues that surround the MDM2 binding site. There is an overall separation in electrostatics of MDM2 and p53 that are mutually complementary and drive association. Upon close approach, there is significant energetic strain as the charges are occluded from water (desolvated). However, the complexation is driven by packing interactions that lead to highly favorable van der Waals interactions. Although the complementarity of the electrostatics of the two surfaces is essential for the two partners to form a complex, steric collisions of Y100 and short ranged van der Waals interactions of F19, W23, L26 of p53 determine the final steps of native complex formation. The electrostatics seem to be evolutionarily conserved, including variations in both partners.














Similar content being viewed by others
References
Naski N et al (2009) Cell Cycle 8:31
Vogelstein B, Lane D, Levine AJ (2000) Nature 408:307
Bottger A et al (1997) Curr Biol 7:860
Moll UM, Petrenko O (2003) Mol Cancer Res 1:1001
Sakurai K, Schubert C, Kahne D (2006) J Am Chem Soc 128:11000
Vassilev LT et al (2004) Science 303:844
Zhong H, Carlson HA (2005) Proteins 58:222
Buolamwini JK, Addo J, Kamath S, Patil S, Mason D, Ores M (2005) Curr Cancer Drug Targets 5:57
Dastidar SG, Lane DP, Verma CS (2008) J Am Chem Soc 130:3514
McCammon JA (2009) Proc Natl Acad Sci USA 106:683
Blundell TL, Fernandez-Recio J (2006) Nature 444:279
Tang C, Iwahara J, Clore GM (2006) Nature 444:383
Sept D, Elcock AH, McCammon JA (1999) J Mol Biol 294:1181
Richter S, Wenzell A, Stein M, Gabdoulline RR, Wade RC (2008) Nucleic Acid Res 36:W276
Heath AP, Kavraki LE, Clementi C (2007) Proteins 68:646
Knotts TA, Rathore N, Schwartz DC, de Pablo JJ (2007) J Chem Phys 126:084901
Macchiarulo A, Giacche N, Carotti A, Baroni M, Cruciani G, Pellicciari R (2008) J Chem Inf Model 48:1999
Dastidar SG, Lane DP, Verma CS, BMC Bioinformatics (in press)
Uhrinova S, Uhrin D, Powers H, Watt K, Zheleva D, Fischer P, McInnes C, Barlow PN (2005) J Mol Biol 350:587
Madhumalar A, Lee HJ, Brown CJ, Lane DP, Verma C (2009) Cell Cycle 8:2828
Feldman-Salit A, Wirtz M, Hell R, Wade RC (2009) J Mol Biol 386:37
Gabdoulline RR, Wade RC (1998) Methods 14:329
Gabdoulline RR, Wade RC (2001) J Mol Biol 306:1139
Elcock AH, Gabdoulline RR, Wade RC, McCammon JA (1999) J Mol Biol 291:149
Lee HJ, Srinivasan D, Coomber D, Lane DP, Verma CS (2007) Cell Cycle 6:2604
Setny P, Geller M (2006) J Chem Phys 125:144717
Gabdoulline RR, Wade RC (2009) J Am Chem Soc 131:9230
Brooks BR et al (2009) J Comp Chem 30:1545
Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP (1996) Science 274:948
Worrall EG, Wawrzynow B, Worrall L, Walkinshaw M, Ball KL, Hupp TR (2009) J Chem Biol 2:113
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) J Chem Phys 79:926
MacKerell AD Jr et al (1998) J Phys Chem B 102:3586
Darden T, York D, Pedersen L (1993) J Chem Phys 98:10089
Ryckaert JP, Ciccotti G, Berendsen HTC (1977) J Comp Phys 23:327
Im W, Lee MS, Brooks CL 3rd (2003) J Comp Chem 24:1691
Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA (2001) Proc Natl Acad Sci USA 98:10037
Thompson JD, Higgins DG, Gibson TJ (1994) Nucleic Acid Res 22:4673
Sali A, Blundell TL (1993) J Mol Biol 234:779
DeLano WL (2002) The Pymol molecular graphics system. DeLano Scientific, San Carlos
Bottger A, Bottger V, Garcia-Echeverria C, Chene P, Hochkeppel HK, Sampson W, Ang K, Howard SF, Picksley SM, Lane DP (1997) J Mol Biol 269:744
Schon O, Friedler A, Bycroft M, Freund SM, Fersht AR (2002) J Mol Biol 323:491
Showalter SA, Bruschweiler-Li L, Johnson E, Zhang F, Bruschweiler R (2008) J Am Chem Soc 130:6472
Shimizu H, Hupp TR (2003) Trends Biochem Sci 28:346
Freedman DA, Epstein CB, Roth JC, Levine AJ (1997) Mol Med 3:248
Czarna A, Popowicz GM, Pecak A, Wolf S, Dubin G, Holak TA (2009) Cell Cycle 8:1176
Popowicz GM, Czarna A, Holak TA (2008) Cell Cycle 7:2441
Hu B, Gilkes DM, Chen J (2007) Cancer Res 67:8810
Massova I, Kollman PA (1999) J Am Chem Soc 121:8133
Van Vlijmen HWT, Karplus M (1999) J Phys Chem B 10:3009
Jones S, Thornton JM (1996) Proc Natl Acad Sci USA 93:13
Acknowledgments
This work was supported by the Biomedical Research Council (Agency for Science, Technology and Research), Singapore. We thank Ivy Law of BII for technical help with Matlab.
Author information
Authors and Affiliations
Corresponding author
Additional information
Dedicated to Professor Sandor Suhai on the occasion of his 65th birthday and published as part of the Suhai Festschrift Issue.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dastidar, S.G., Madhumalar, A., Fuentes, G. et al. Forces mediating protein–protein interactions: a computational study of p53 “approaching” MDM2. Theor Chem Acc 125, 621–635 (2010). https://doi.org/10.1007/s00214-009-0682-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00214-009-0682-1